No Yes
0.9667 0.0333 Generative Models
MSSC 6250 Statistical Machine Learning
Department of Mathematical and Statistical Sciences
Marquette University
Background: Bayes Classifier
Bayes Classifier
The test error rate is minimized, on average, by a classifier that assigns each observation to the most likely category, given its predictor values.
This fact leads to
Bayes (Optimal) Classifier: Assign a test observation \(y_j\) with predictor \(x_j\) to class \(k\) for which the conditional probability \(P(Y_j = k \mid X = x_j)\) is largest.
The Bayes classifier produces the lowest possible test error rate, called Bayes error rate.
The problem is
We never know \(P(Y_j = k \mid X = x_j)\) 😭
The goal of soft classifiers is to estimate it, like logistic regression:
\[ \widehat{\pi}(\mathbf{x}) = \widehat{P}(Y = 1 \mid \mathbf{x}) = \frac{1}{1+e^{-\mathbf{x}'\widehat{\boldsymbol{\beta}}}}\]
Generative Models
What is a Generative Model?
- So far predictors are assumed fixed and known, and we care about the conditional distribution, \(p(y \mid \mathbf{x})\)
- Logistic regression (LR)
- A generative model considers the joint distribution of the response and predictors, i.e., \(p(y, \mathbf{x})\)
- Discriminant analysis
- Naive Bayes (NB)
Why not just logistic regression?
When the classes are substaintially separated, the parameter estimates for the logistic regression are unstable.
If the distribution of the predictors is approximately normal in each of the classes and the sample size is small, the generative models could perform better and more stable.
Generative Modeling
- To infer/predict \(p(y = k \mid \mathbf{x})\) from a generative model, Bayes theorem comes into play:
\[\begin{align} p(y = k \mid \mathbf{x}) &= \frac{p(y = k, \mathbf{x})}{p(\mathbf{x})} = \frac{p(y = k)p(\mathbf{x}\mid y = k) }{p(\mathbf{x})} = \frac{p(y = k)p(\mathbf{x}\mid y = k)}{\sum_{l=1}^K p(y = k)p(\mathbf{x}\mid y = l)}\\ &=\frac{\pi_kf_k(\mathbf{x})}{\sum_{l=1}^K\pi_lf_l(\mathbf{x})}\end{align}\]
- To get the posterior \(p(y = k \mid \mathbf{x})\), we need
- prior \(\pi_k = p(y = k)\)
- likelihood \(f_k(\mathbf{x}) = p(\mathbf{x}\mid y = k)\)
Note
Different generative models use different priors and likelihoods, resulting in different strengths and weakness.
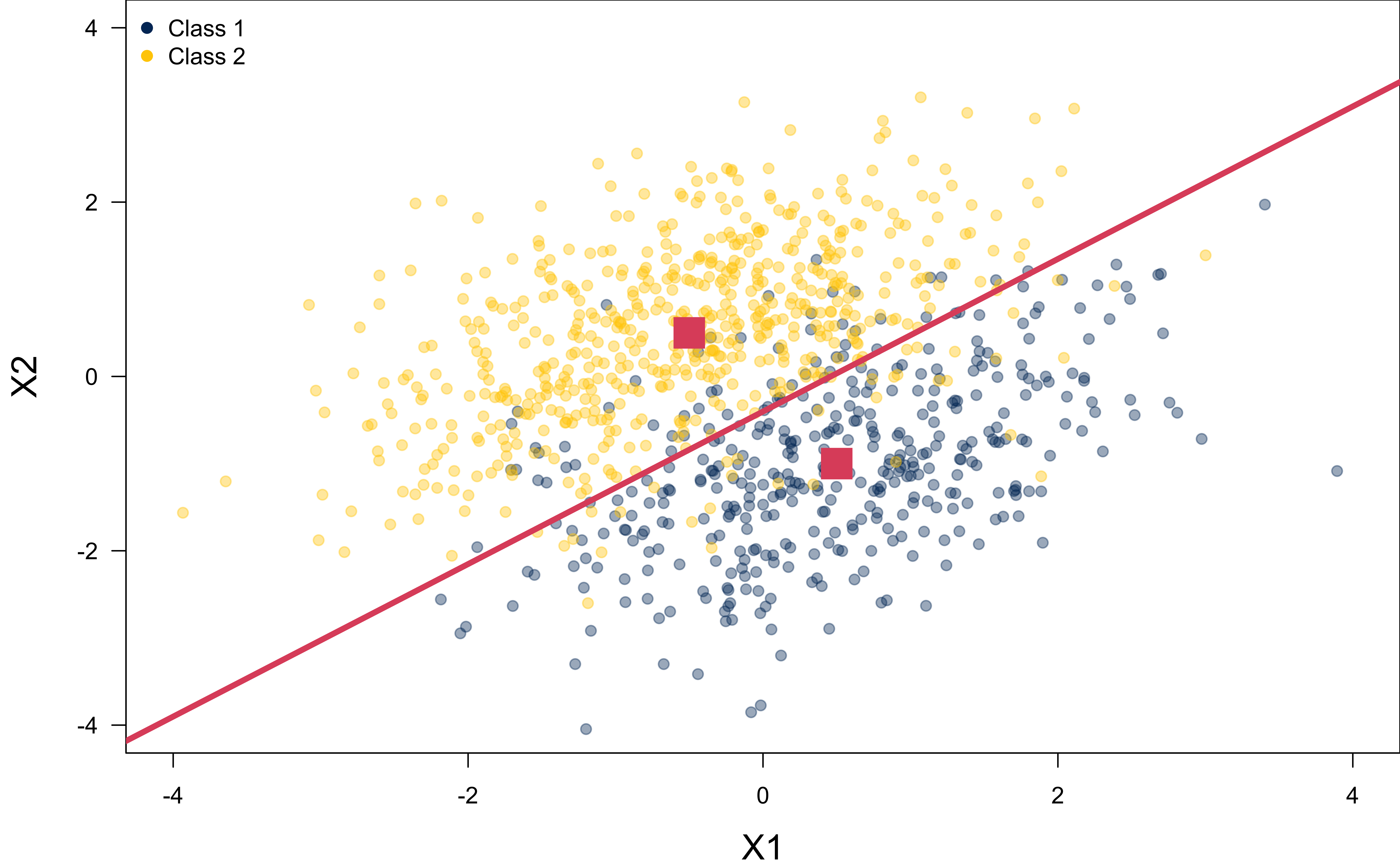
Bayes Decision Boundary
How do we find/estimate the Bayes decision boundary generated by the Bayes classifier?
Linear Discriminant Analysis (LDA)
\[\begin{align} p(y = k \mid \mathbf{x}) = \frac{p(y = k)p(\mathbf{x}\mid y = k)}{\sum_{l=1}^K p(y = k)p(\mathbf{x}\mid y = l)} =\frac{\pi_kf_k(\mathbf{x})}{\sum_{l=1}^K\pi_lf_l(\mathbf{x})}\end{align}\]
- Estimate the prior \(\pi_k = p(y = k)\) using the proportion of the training labels that belong to the \(k\)th class: \[\hat{\pi}_k = n_k / n \]
-
LDA assumption for \(f_k(\mathbf{x})\): \(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma \right)}\)
- \(\boldsymbol{\mu}_k\): class-specific mean vector
- \(\Sigma\): covariance matrix common to all \(K\) classes
Two Multivariate Normals with Common Covariance
\(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma \right)}\)
\(\boldsymbol{\mu}_1 = (0.5, -1)'\), \(\boldsymbol{\mu}_2 = (-0.5, 0.5)'\)
\(\Sigma = \begin{pmatrix} 1 & 0.5 \\ 0.5 & 1 \end{pmatrix}\)
\(\pi_1 = 0.4\), \(\pi_2 = 0.6\)
Each class has its own predictor mean.
The predictor’s variability are the same for the two classes.
Parameter Estimation for LDA
\(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma \right)}\)
- Estimating \(p(\mathbf{x}\mid y = k)\) is reduced to estimating \(\boldsymbol{\mu}_1, \dots, \boldsymbol{\mu}_K\) and \(\Sigma\).
- Centroid/sample mean: \(\widehat{\boldsymbol{\mu}}_k = \frac{1}{n_k} \sum_{i: \,y_i = k} \mathbf{x}_i\)
- Pooled covariance matrix: \[\widehat \Sigma = \frac{1}{n-K} \sum_{k=1}^K \sum_{i : \, y_i = k} (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k) (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k)' = \sum_{k=1}^K \frac{n_k-1}{n-K}\widehat \Sigma_k\] where \(\widehat \Sigma_k = \frac{1}{n_k-1}\sum_{i : \, y_i = k} (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k) (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k)'\)
mu1_hat =[1] 0.47 -1.01mu2_hat =[1] -0.54 0.51Sigma_hat = [,1] [,2]
[1,] 1.13 0.54
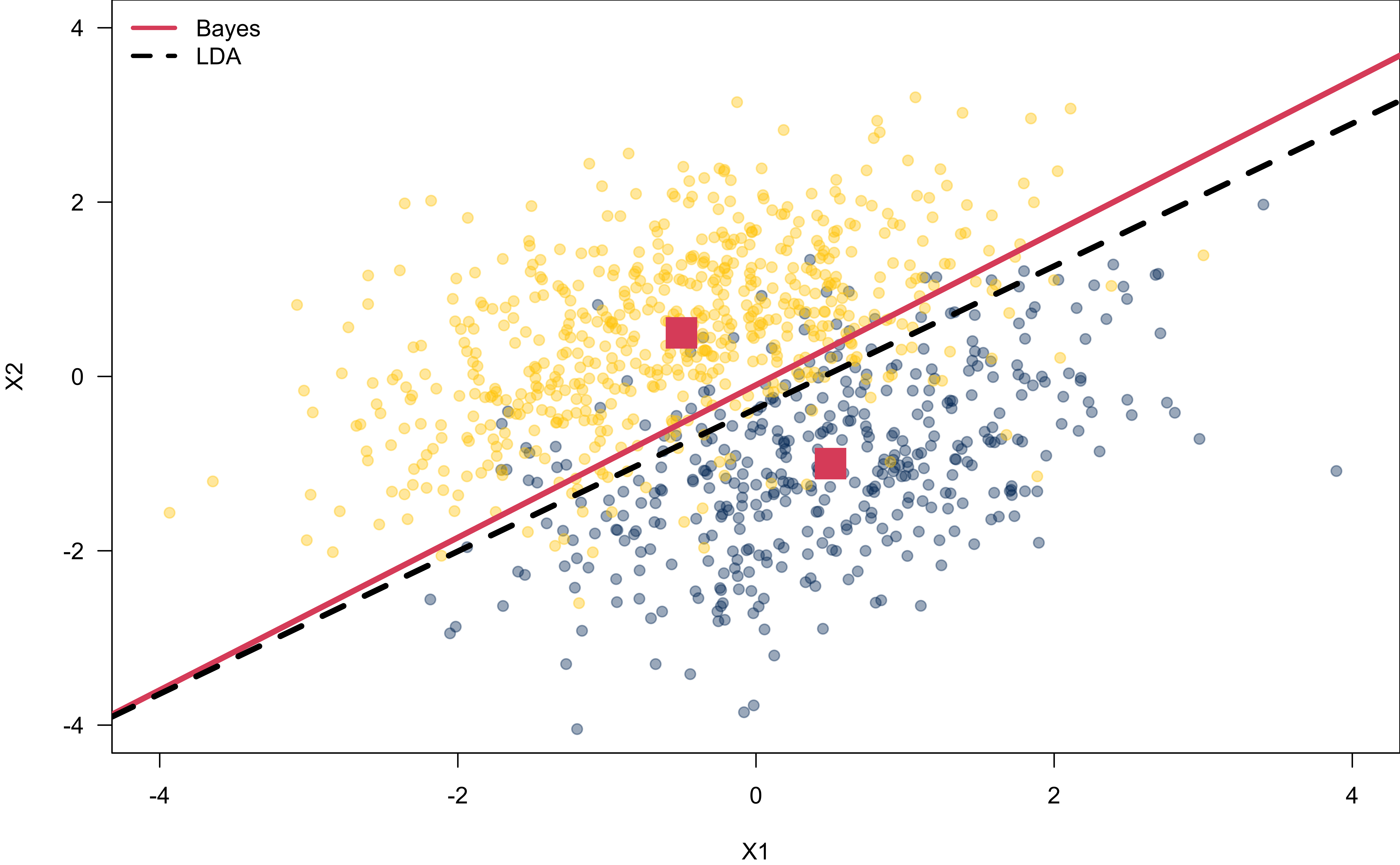
[2,] 0.54 1.02Decision Boundary by LDA
Discriminant Function
The likelihood is Gaussian
\[f_k(x) = \frac{1}{(2\pi)^{p/2} |\Sigma|^{1/2}} \exp\left[ -\frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) \right]\]
The log-likelihood is
\[\begin{align} \log f_k(\mathbf{x}) =&~ -\log \big((2\pi)^{p/2} |\Sigma|^{1/2} \big) - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)'\Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) \\ =&~ - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) + \text{Constant} \end{align}\]
The maximum a posteriori estimate is
\[\begin{align} \widehat f(\mathbf{x}) =& ~\underset{k}{\arg\max} \,\, \log \big( \pi_k f_k(\mathbf{x}) \big) \\ =& ~\underset{k}{\arg\max} \,\, - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) + \log(\pi_k) \end{align}\]
Discriminant Function
\[\begin{align} \widehat f(\mathbf{x}) =& ~\underset{k}{\arg\max} \,\, - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) + \log(\pi_k) \end{align}\]
- \((\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k)\) is the Mahalanobis distance between \(\mathbf{x}\) and the centroid \(\boldsymbol{\mu}_k\).
- Classify \(y\) so that its \(\mathbf{x}\) and the centroid of the labeled class is the closest (after adjusting the for prior). \[\begin{align} & - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) + \log(\pi_k)\\ &=~ \mathbf{x}' \underbrace{\Sigma^{-1} \boldsymbol{\mu}_k}_{\mathbf{w}_k} \underbrace{- \frac{1}{2}\boldsymbol{\mu}_k' \Sigma^{-1} \boldsymbol{\mu}_k + \log(\pi_k)}_{b_k} + \text{const of } k \\ &=~ \mathbf{x}' \mathbf{w}_k + b_k + \text{const of } k\\ &=~ \delta_k(\mathbf{x}) + \text{const of } k \end{align}\] where \(\delta_k(\mathbf{x})\) the discriminant function which is linear in \(\mathbf{x}\).
Discriminant Function
The decision boundary of LDA is a linear function of \(\mathbf{x}\).
The boundary between two classes \(k\) and \(l\) is where they have the same density value.
\[\begin{align} \delta_k(\mathbf{x}) &= \delta_l(\mathbf{x}) \\ \Leftrightarrow \quad \mathbf{x}' \mathbf{w}_k + b_k &= \mathbf{x}' \mathbf{w}_l + b_l \\ \Leftrightarrow \quad \mathbf{x}' (\mathbf{w}_k - \mathbf{w}_l) + (b_k - b_l) &= 0. \\ \end{align}\]
\[\begin{align} &\delta_k(\mathbf{x}) - \delta_l(\mathbf{x}) \\ \propto & \quad \log \left(\pi_k f_k(\mathbf{x})\right) - \log \left(\pi_l f_l(\mathbf{x}))\right)\\ \propto & \quad \log \left(P(Y = k \mid \mathbf{x})\right) - \log \left(P(Y = l \mid \mathbf{x})\right) \\ \propto & \quad \log \left( \frac{P(Y = k \mid \mathbf{x})}{P(Y = l \mid \mathbf{x})}\right) \end{align}\]
Interpretation of LDA
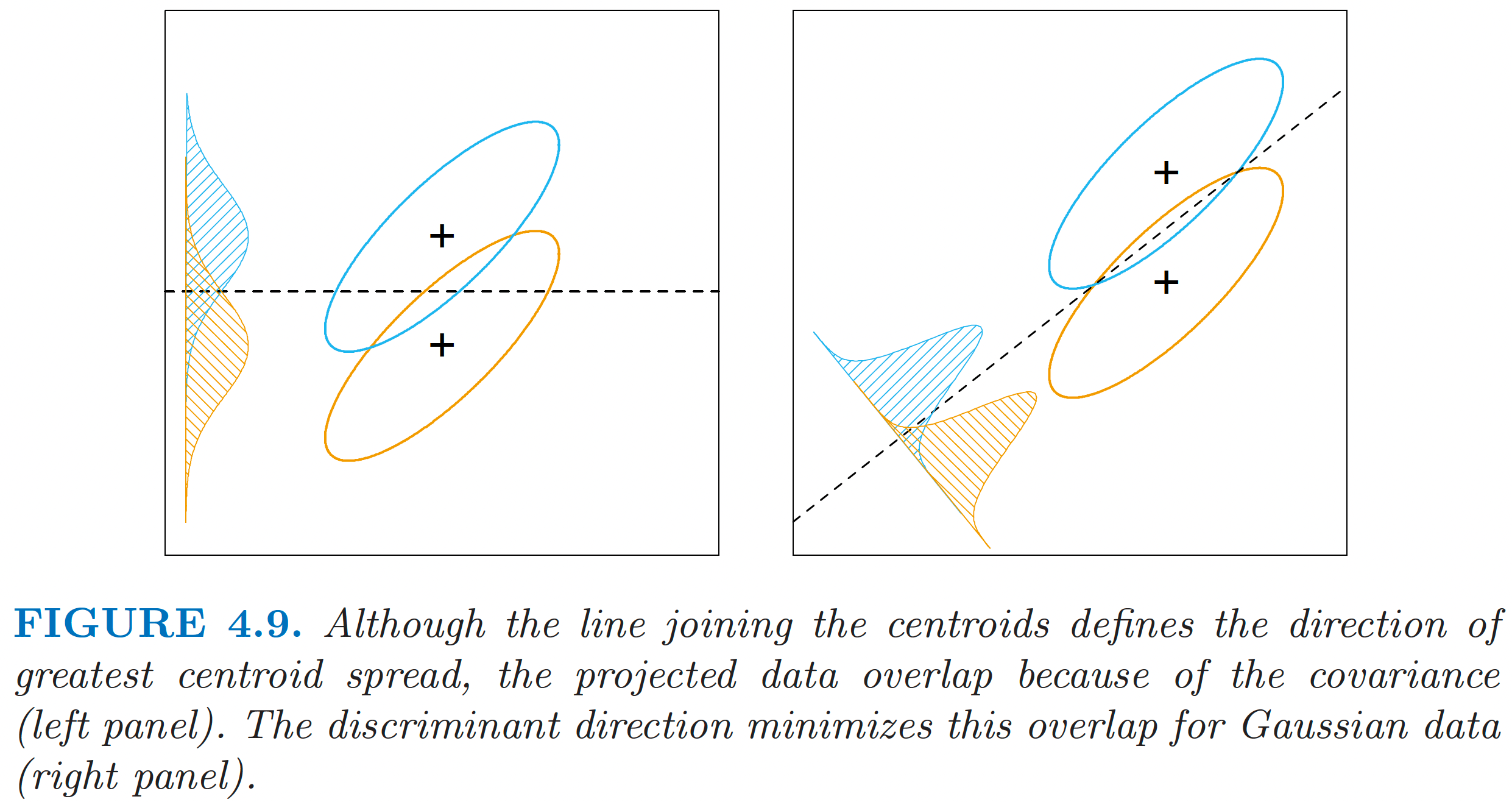
ESL Fig 4.9
MASS::lda()
LDA
lda_fit <- MASS::lda(default ~ balance, data = Default)
lda_pred <- predict(lda_fit, data = Default)
(lda_conf <- table(lda_pred$class, Default$default))
No Yes
No 9643 257
Yes 24 76- training accuracy is 0.972
- sensitivity is 0.228
Logistic Regression
No Yes
FALSE 9625 233
TRUE 42 100- training accuracy is 0.973
- sensitivity is 0.3
Quadratic Discriminant Analysis (QDA)
-
LDA assumption for \(f_k(\mathbf{x})\): \(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma \right)}\)
- \(\Sigma\): covariance matrix common to all K classes
-
QDA assumption for \(f_k(\mathbf{x})\): \(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma_k \right)}\)
- \(\Sigma_k\): each class has its own covariance matrix
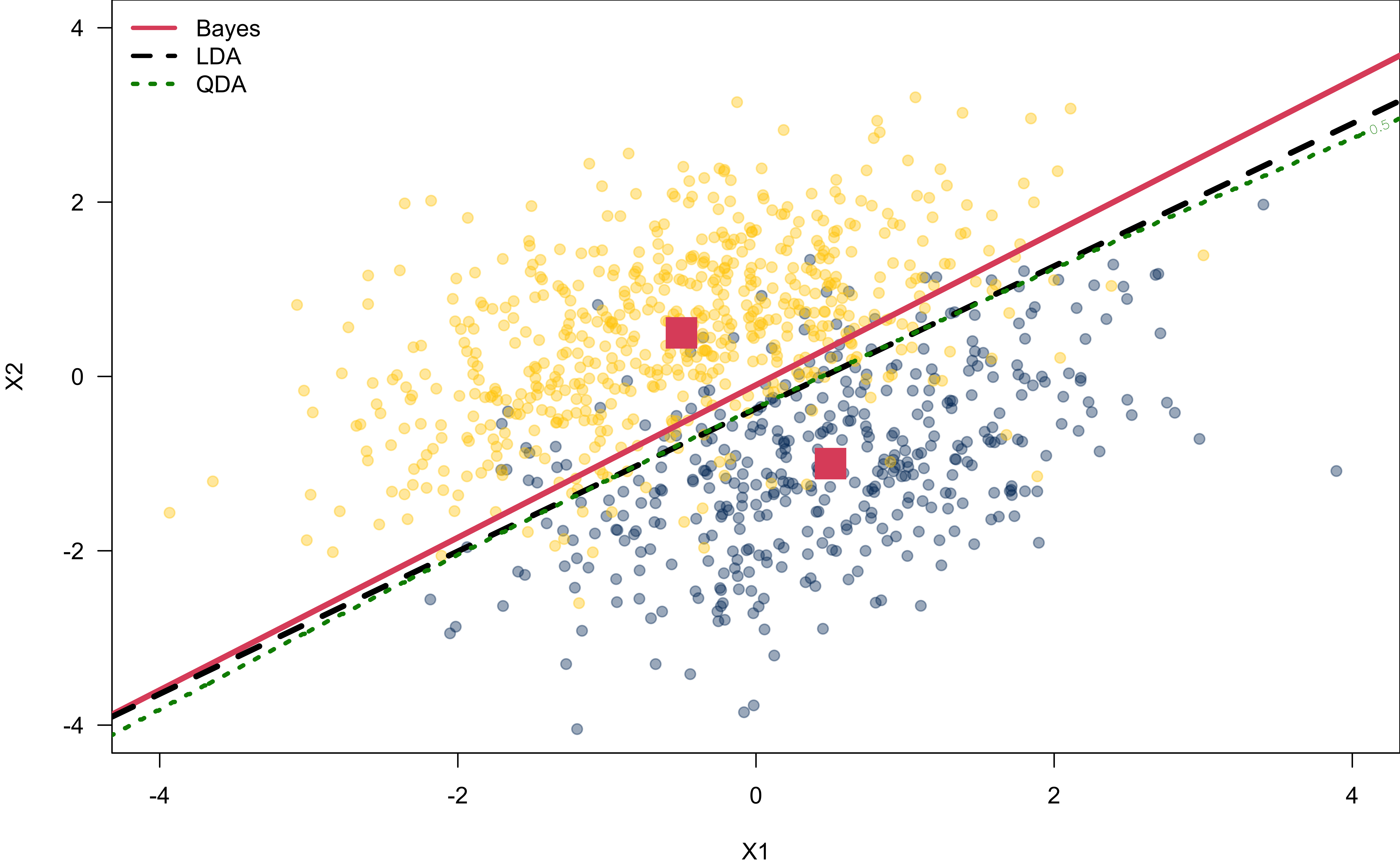
The discriminant function for QDA is
\[\begin{align} \delta_k(\mathbf{x}) &= - \frac{1}{2} (\mathbf{x}- \boldsymbol{\mu}_k)' \Sigma_k^{-1} (\mathbf{x}- \boldsymbol{\mu}_k) - \frac{1}{2} \log |\Sigma_k |+ \log(\pi_k) \\ &= \mathbf{x}' \mathbf{W}_k \mathbf{x}+ \mathbf{x}'\mathbf{w}_k + b_k \end{align}\] which is a quadratic function of \(\mathbf{x}\).
The boundary between class \(k\) and \(l\) for QDA is
\[\begin{align} \mathbf{x}' (\mathbf{W}_k - \mathbf{W}_l) \mathbf{x}+ \mathbf{x}' (\mathbf{w}_k - \mathbf{w}_l) + (b_k - b_l) = 0\\ \end{align}\]
Parameter Estimation for QDA
The estimation procedure is almost the same as LDA.
\(\hat{\pi}_k = n_k / n\)
Centroids: \(\widehat{\boldsymbol{\mu}}_k = \frac{1}{n_k} \sum_{i: \,y_i = k} \mathbf{x}_i\)
Individual covariance matrix: \[\widehat \Sigma_k = \frac{1}{n_k-1}\sum_{i : \, y_i = k} (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k) (\mathbf{x}_i - \widehat{\boldsymbol{\mu}}_k)'\]
Two Multivariate Normals with Different Covariance Matrices
\(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma_k \right)}\)
\(\boldsymbol{\mu}_1 = (0.5, -1)'\)
\(\boldsymbol{\mu}_2 = (-0.5, 0.5)'\)
\(\Sigma_1 = \begin{pmatrix} 1 & 0.5 \\ 0.5 & 1 \end{pmatrix}\)
\(\Sigma_2 = \begin{pmatrix} 1 & -0.5 \\ -0.5 & 1 \end{pmatrix}\)
\(\pi_1 = 0.4\), \(\pi_2 = 0.6\)
Each class has its own predictor mean and covariance matrix.
Decision Boundary by QDA
MASS::qda
LDA
No Yes
No 9643 257
Yes 24 76QDA
qda_fit <- MASS::qda(default ~ balance, data = Default)
qda_pred <- predict(qda_fit, data = Default)
(qda_conf <- table(qda_pred$class, Default$default))
No Yes
No 9639 246
Yes 28 87- training accuracy is 0.973
- sensitivity is 0.261
Naive Bayes (NB)
-
LDA assumption for \(f_k(\mathbf{x})\): \(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma \right)}\)
- \(\Sigma\): covariance matrix common to all K classes
-
QDA assumption for \(f_k(\mathbf{x})\): \(\color{blue}{(\mathbf{x}\mid y = k) \sim N_p\left(\boldsymbol{\mu}_k, \Sigma_k \right)}\)
- \(\Sigma_k\): each class has its own covariance matrix
-
NB assumption for \(f_k(\mathbf{x})\):
- features within each class are independent!
- \(f_k(\mathbf{x}) = f_{k1}(x_1) \times f_{k2}(x_2) \times \cdots \times f_{kp}(x_p)\)
Considering the joint distribution of predictors is a pain when \(p\) is large.
We don’t believe \(p\) predictors are not associated at all, but this “naive” assumption leads to decent results when \(n\) is not large enough relative to \(p\).
The naive independence assumption introduces some bias, but reduces variance.
\[\begin{align} p(y = k \mid \mathbf{x}) =\frac{\pi_kf_k(\mathbf{x})}{\sum_{l=1}^K\pi_lf_l(\mathbf{x})} = \frac{\pi_k\prod_{j=1}^p f_{kj(x_j)}}{\sum_{l=1}^K\pi_l\prod_{j=1}^p f_{lj(x_j)}} \end{align}\]
Options for Estimating \(f_{kj}(x_j)\)
- Numerical \(X_j\)
- Parametric: \[(X_j \mid Y = k) \sim N\left(\mu_{jk}, \sigma_{jk}^2 \right).\] It’s QDA with additional assumption that \(\Sigma_k\) is diagonal.
- Non-parametric: Estimate \(f_{kj}(x_j)\) by (discrete) histogram, (continuous) kernel density estimator, etc.
-
Categorical \(X_j\)
- \(f_{kj}(x_j) \approx\) the proportion of training data for \(x_j\) corresponding to class \(k\).
Class 1
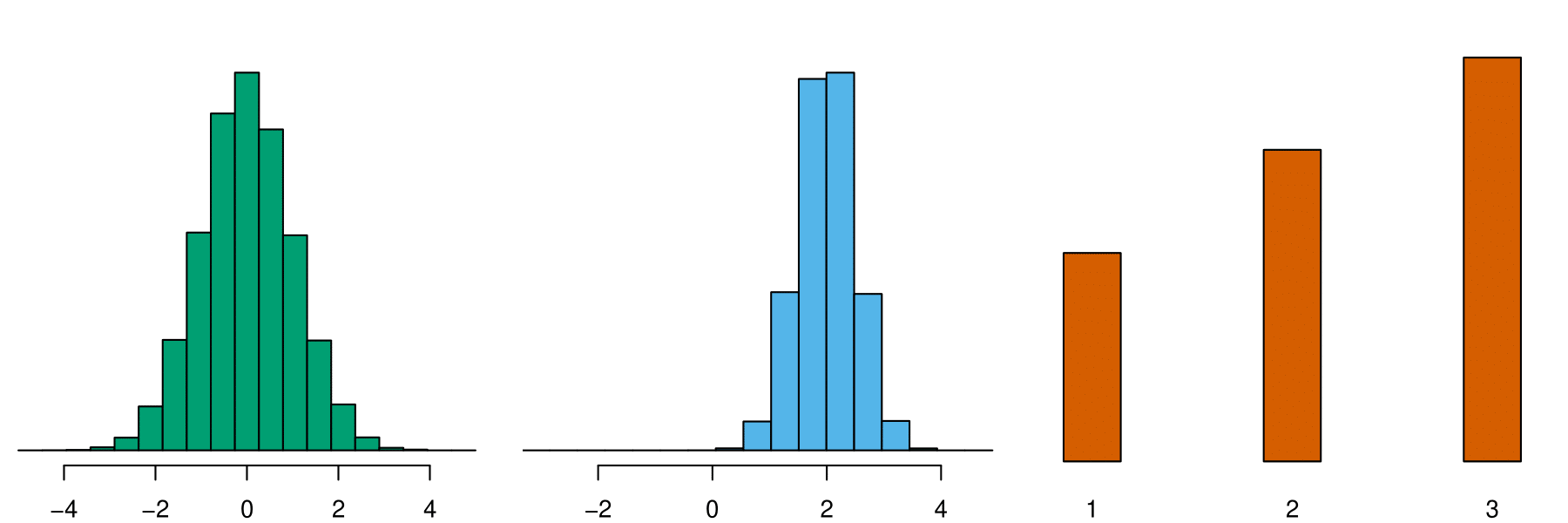
Class 2
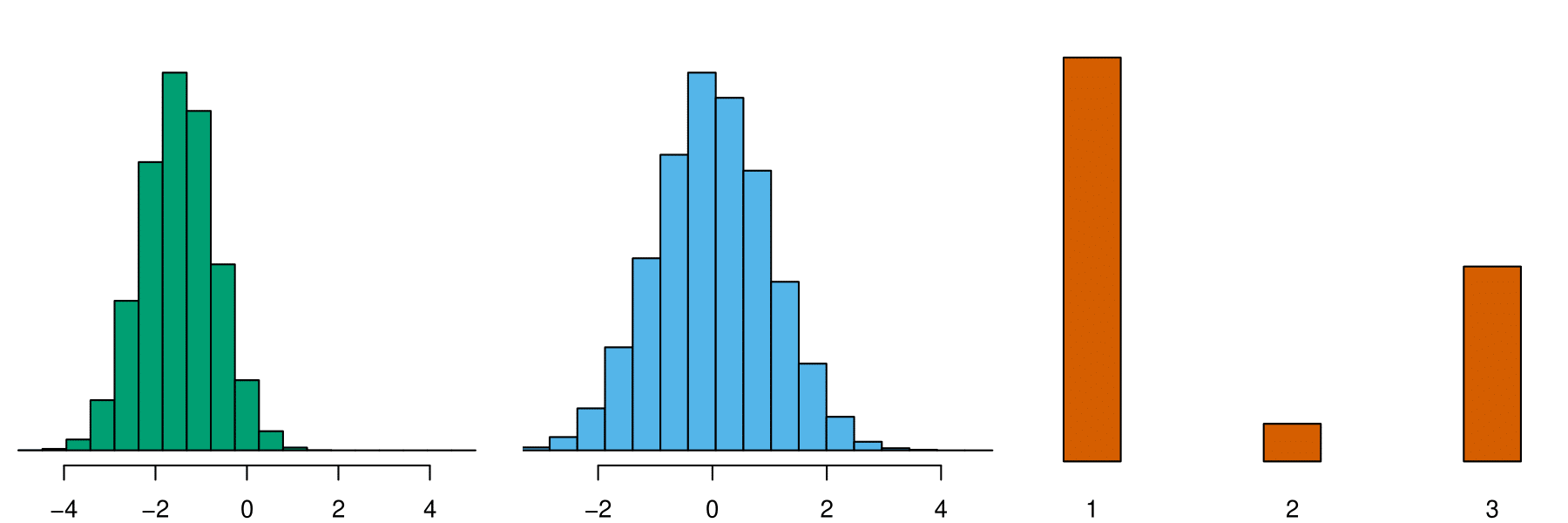
e1071::naiveBayes()
QDA
No Yes
No 9639 246
Yes 28 87Naive Bayes - By default, each feature is Gaussian distributed.
nb_fit <- e1071::naiveBayes(default ~ balance, data = Default)
nb_pred <- predict(nb_fit, Default)
(nb_conf <- table(nb_pred, Default$default))
nb_pred No Yes
No 9639 246
Yes 28 87- training accuracy is 0.973
- sensitivity is 0.261
Comparison of Classification Methods (LR, LDA, QDA, NB)
Log Odds of LR and LDA
Suppose class \(K\) is the baseline.
- LR
\[\log \left( \frac{P(Y = k \mid \mathbf{x})}{P(Y = K \mid \mathbf{x})}\right) = \beta_{k0} + \beta_{k1}x_{1} + \dots + \beta_{kp}x_{p}\]
- LDA
\[\log \left( \frac{P(Y = k \mid \mathbf{x})}{P(Y = K \mid \mathbf{x})}\right) = a_k + \sum_{j=1}^pc_{kj}x_j\] where \(a_k = \log(\pi_k / \pi_K) - \frac{1}{2}(\boldsymbol{\mu}_k + \boldsymbol{\mu}_K)'\Sigma^{-1}(\boldsymbol{\mu}_k - \boldsymbol{\mu}_K)\) and \(c_{kj}\) is the \(j\)th element of \(\Sigma^{-1}(\boldsymbol{\mu}_k - \boldsymbol{\mu}_K)\).
Both log odds are linear in \(\mathbf{x}\).
Log Odds of QDA and NB
- QDA
\[\log \left( \frac{P(Y = k \mid \mathbf{x})}{P(Y = K \mid \mathbf{x})}\right) = a_k + \sum_{j=1}^pc_{kj}x_j + \sum_{j=1}^p\sum_{l=1}^p d_{kjl}x_jx_l\] The log odds is quadratic in \(\mathbf{x}\), where \(a_k\), \(c_{kj}\) and \(d_{kjl}\) are functions of \(\pi_k\), \(\pi_K\), \(\boldsymbol{\mu}_k\), \(\boldsymbol{\mu}_K\), \(\Sigma_k\) and \(\Sigma_K\).
- NB
\[\log \left( \frac{P(Y = k \mid \mathbf{x})}{P(Y = K \mid \mathbf{x})}\right) = a_k + \sum_{j=1}^pg_{kj}(x_j)\] where \(a_k = \log(\pi_k / \pi_K)\) and \(g_{kj}(x_j) = \log\left( \frac{f_{kj}(x_j)}{f_{Kj}(x_j)}\right)\).
The log odds takes the form of a generalized additive model!
Comparison of Log Odds
LDA is a special case of QDA with \(d_{kjl} = 0\) for all \(k, j\) and \(l\). \((\Sigma_1 = \cdots = \Sigma_k = \Sigma)\)
-
Any classifier with a linear decision boundary is a special case of NB with \(g_{kj}(x_j) = c_{kj}x_j.\)
- LDA is a special case of NB. Really !? 😲
NB is a special case of LDA if \(f_{kj}(x_j) = N\left(\mu_{jk}, \sigma_{j}^2 \right)\). \((\Sigma = \text{diag}(\sigma_1^2, \dots, \sigma_p^2))\)
-
Neither QDA nor NB is a special case of the other
- NB is more
- flexible for any transformation of \(x_j\), \(g_{kj}(x_j)\).
- restrictive because of its pure additive fit with no quadratic interactions \(d_{kjl}x_jx_l\).
- NB is more
Which Method to Use
None of these methods uniformly dominates the others.
-
The choice of method depends on
- the true distribution of \(X_j\)s in each class
- the sizes of \(n\) and \(p\) that controls bias-variance trade-off
LDA outperforms LR when predictors are approximately normal, but use LR when inputs are away from normal.
Use LDA but not QDA when variability of predictors are similar among all \(K\) classes.
Do not use naive Bayes when predictors are clearly correlated.
Use QDA when predictors clearly have different covariance structure for each class.
Which Method to Use
QDA is more flexible than LDA (more parameters to be estimated), and hence low bias and high variance.
LDA tends to be better than QDA if \(n\) is relatively small, and so reducing variance is crucial.
QDA is recommended if \(n\) is relatively large so that the variance of the classifier is not a major concern.
Naive Bayes works when \(n\) is not large enough relative to \(p\).