Gaussian Processes ♾️
MSSC 6250 Statistical Machine Learning
Department of Mathematical and Statistical Sciences
Marquette University
Gaussian Variables
Univariate Guassian
- \(Y \sim N(\mu, \sigma^2)\)

Multivariate Guassian
“Multivariate” \(=\) two or more random variables
\(\mathbf{Y}\in \mathbb{R}^d \sim N_d\left(\boldsymbol \mu, \boldsymbol \Sigma\right)\)
- Bivariate Gaussian (\(d=2\)):
\[\mathbf{Y}= \begin{pmatrix} Y_1 \\ Y_2 \end{pmatrix}\] \[\boldsymbol \mu= \begin{pmatrix} \mu_1 \\ \mu_2 \end{pmatrix}\] \[\boldsymbol \Sigma= \begin{pmatrix} \sigma_1^2 & \rho_{12}\sigma_1\sigma_2\\ \rho_{12}\sigma_1\sigma_2 & \sigma_2^2 \end{pmatrix}\]
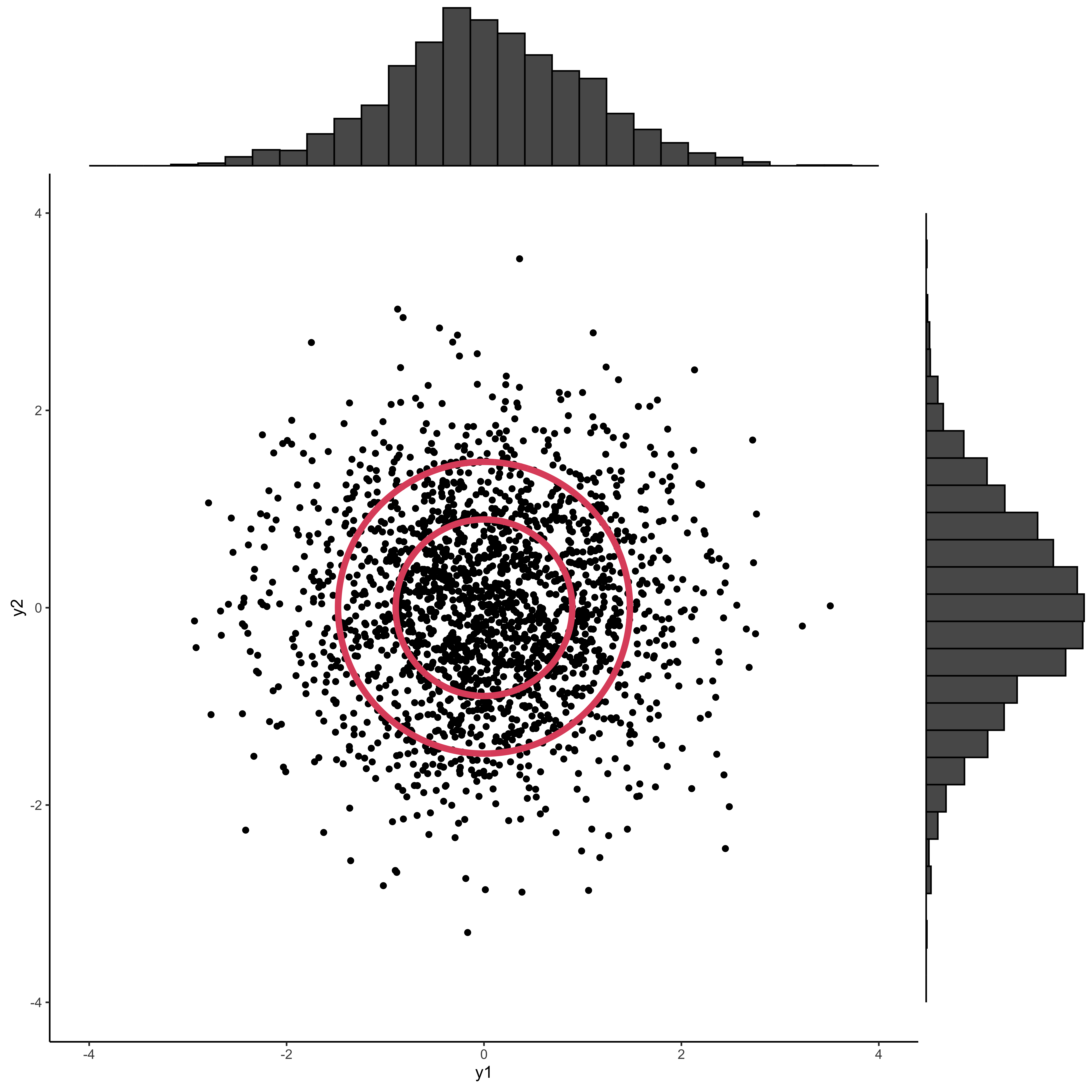
\[\boldsymbol \mu= \begin{pmatrix} 0 \\ 0 \end{pmatrix}\] \[\boldsymbol \Sigma= \begin{pmatrix}1 & 0\\ 0 & 1 \end{pmatrix}\]
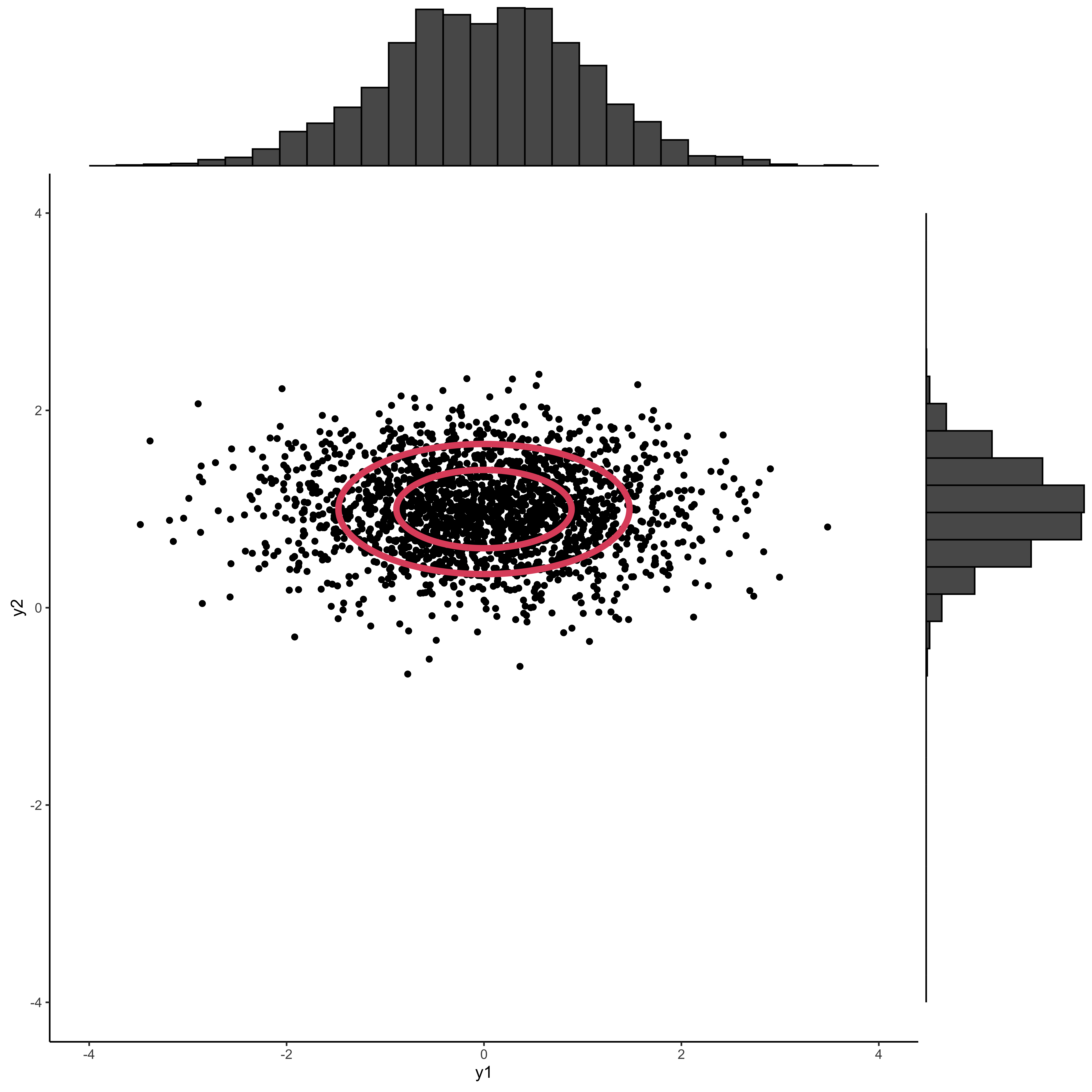
\[\boldsymbol \mu= \begin{pmatrix} 0 \\ 1 \end{pmatrix}\] \[\boldsymbol \Sigma= \begin{pmatrix}1 & 0\\ 0 & 0.2 \end{pmatrix}\]
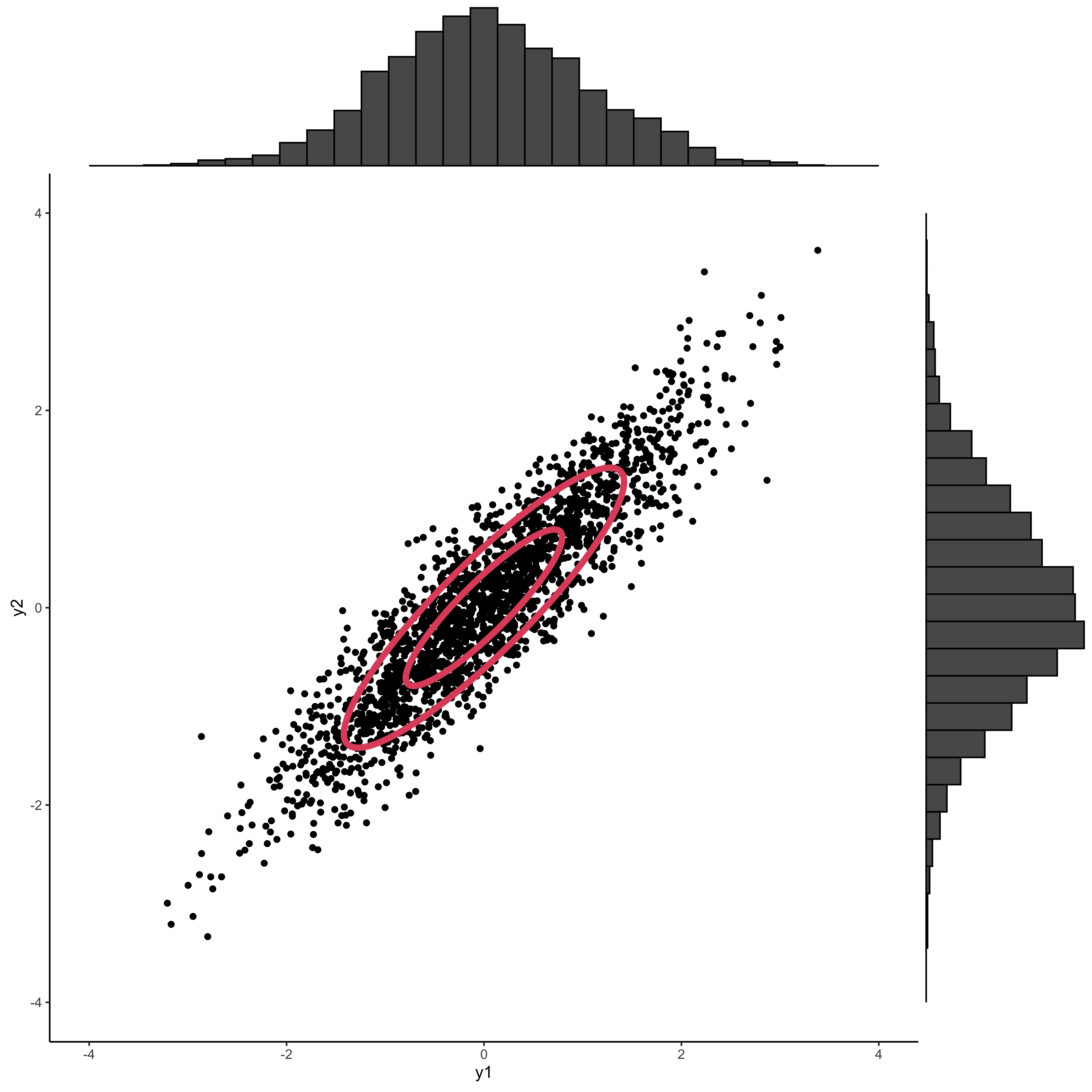
\[\boldsymbol \mu= \begin{pmatrix} 0 \\ 0 \end{pmatrix}\] \[\boldsymbol \Sigma= \begin{pmatrix}1 & 0.9\\ 0.9 & 1 \end{pmatrix}\]
Three or More Variables
- Hard to visualize in dimensions \(> 2\), so stack points next to each other.

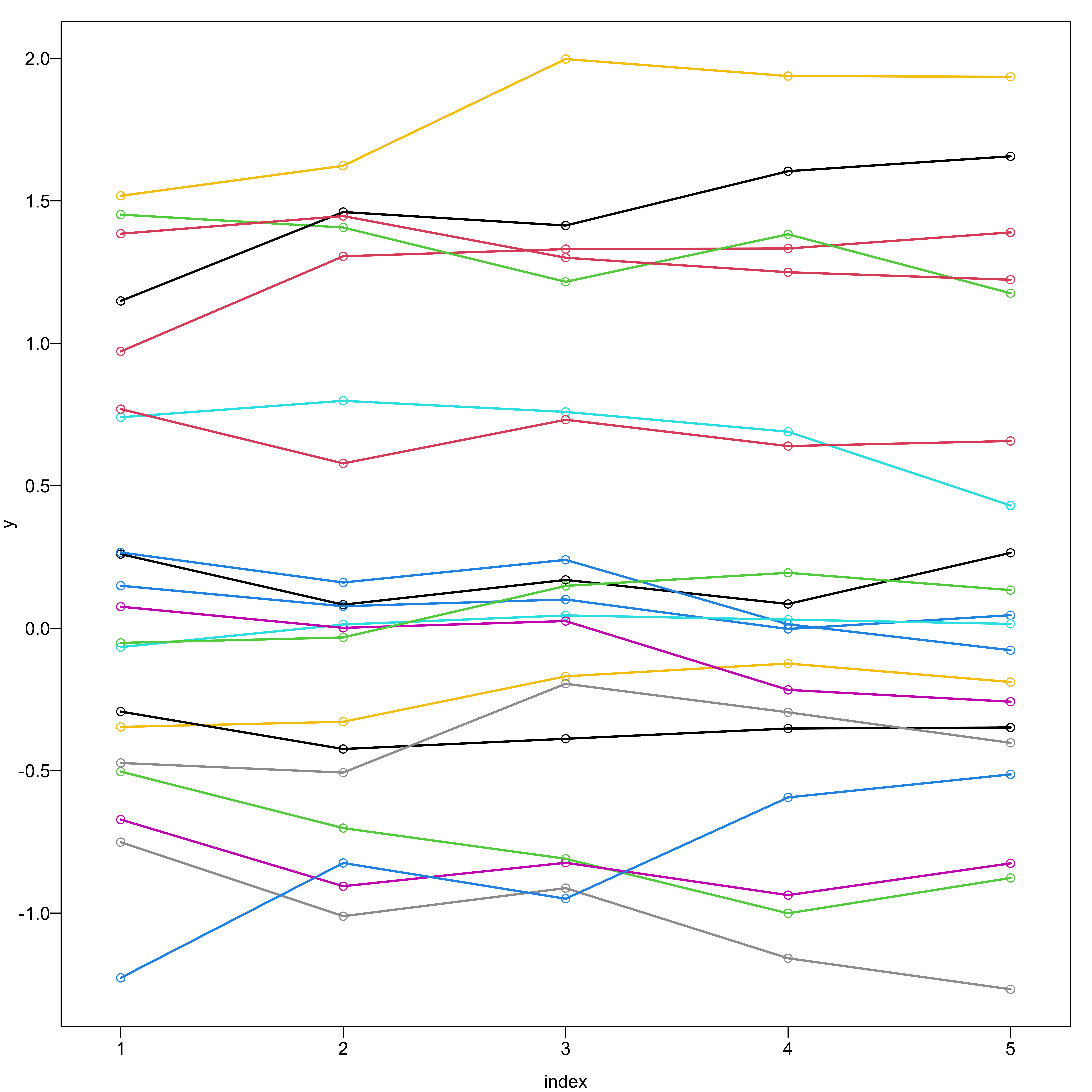
\(d = 5\)
\[\boldsymbol \mu= \begin{pmatrix} 0 \\ 0 \\ 0\\0\\0\end{pmatrix} \quad \quad \boldsymbol \Sigma= \begin{pmatrix}1 & 0.99 & 0.98 & 0.97 & 0.96\\ 0.99 & 1 & 0.99 & 0.98 & 0.97 \\ 0.98 & 0.99 & 1 & 0.99 & 0.98 \\ 0.97 & 0.98 & 0.97 & 1 & 0.99\\0.96 & 0.97 & 0.98 & 0.99 & 1 \end{pmatrix}\]
- Each line is one sample (path).
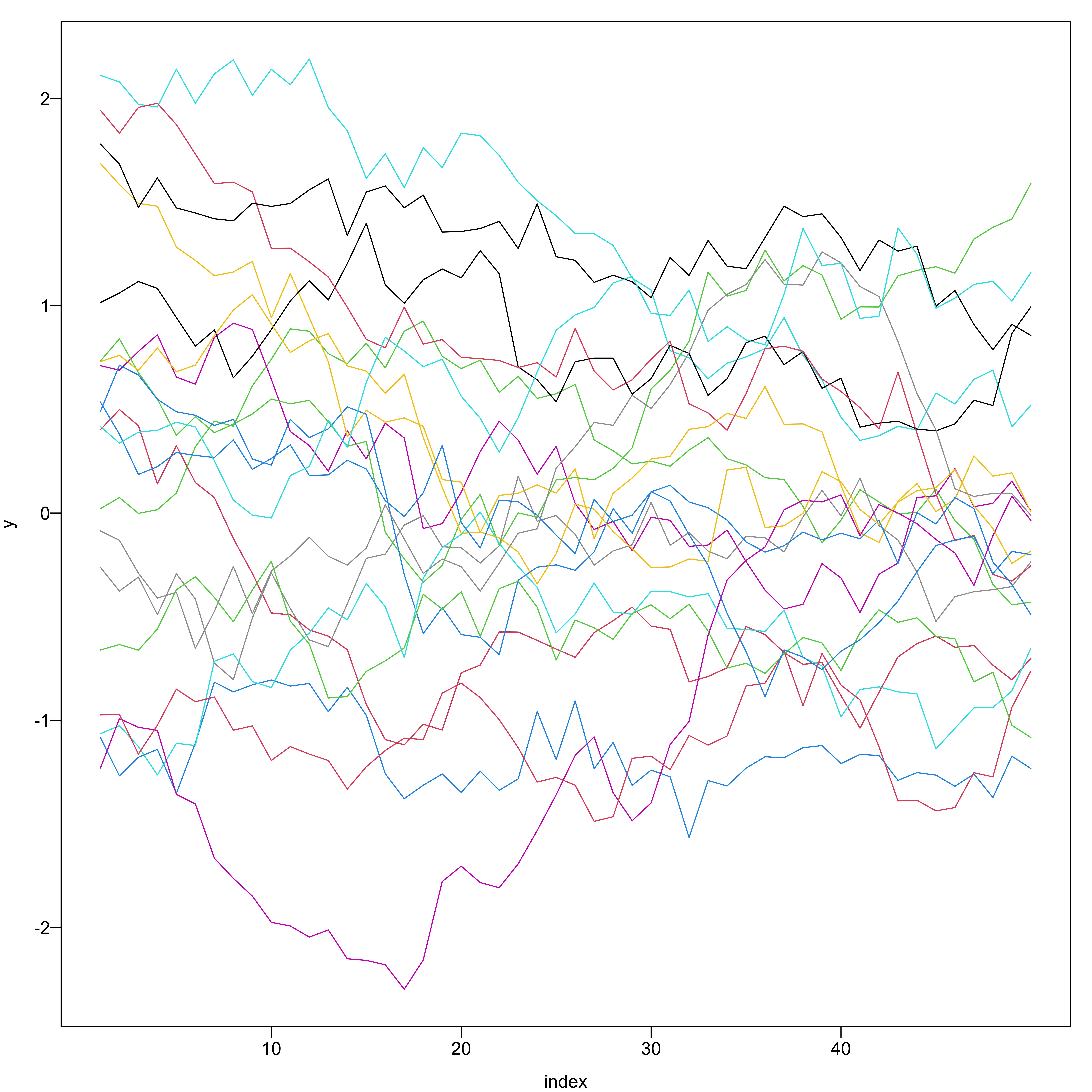
\(d = 50\)
\[\boldsymbol \mu= \begin{pmatrix} 0 \\ 0 \\ \vdots \\0\\0\end{pmatrix} \quad \quad \boldsymbol \Sigma= \begin{pmatrix} 1 & 0.99 & 0.98 & 0.97 & 0.96 & \cdots \\ 0.99 & 1 & 0.99 & 0.98 & 0.97 & \cdots\\ 0.98 & 0.99 & 1 & 0.99 & 0.98 & \cdots\\ 0.97 & 0.98 & 0.97 & 1 & 0.99 & \cdots\\ 0.96 & 0.97 & 0.98 & 0.99 & 1 & \cdots \\ \vdots & \vdots & \vdots & \vdots & \vdots & \vdots & \end{pmatrix}\]
Each line is one sample (path).
-
Think of Gaussian processes as an infinite dimensional distribution over functions
- all we need to do is change notation
Gaussian Processes
Gaussian Processes
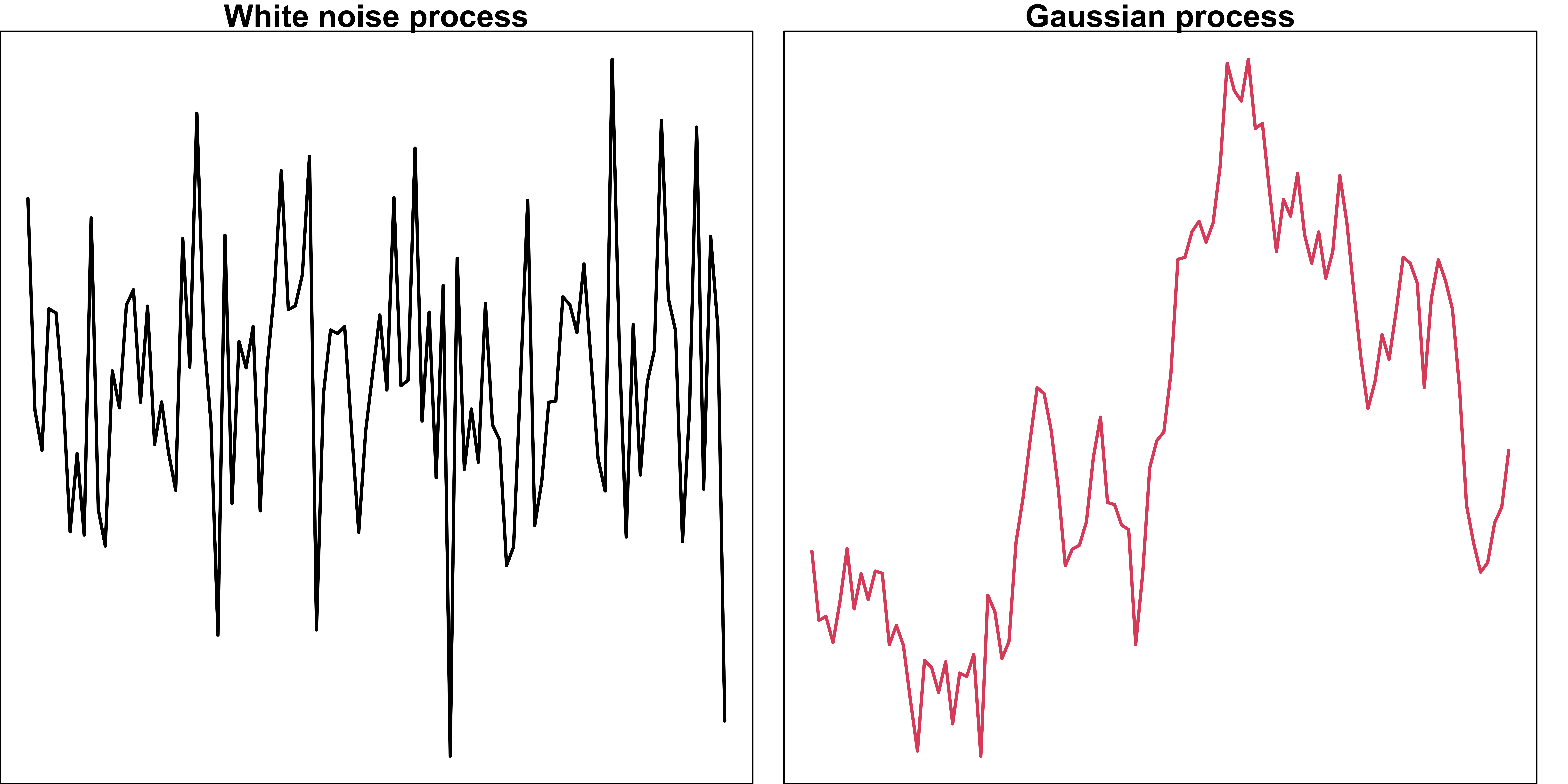
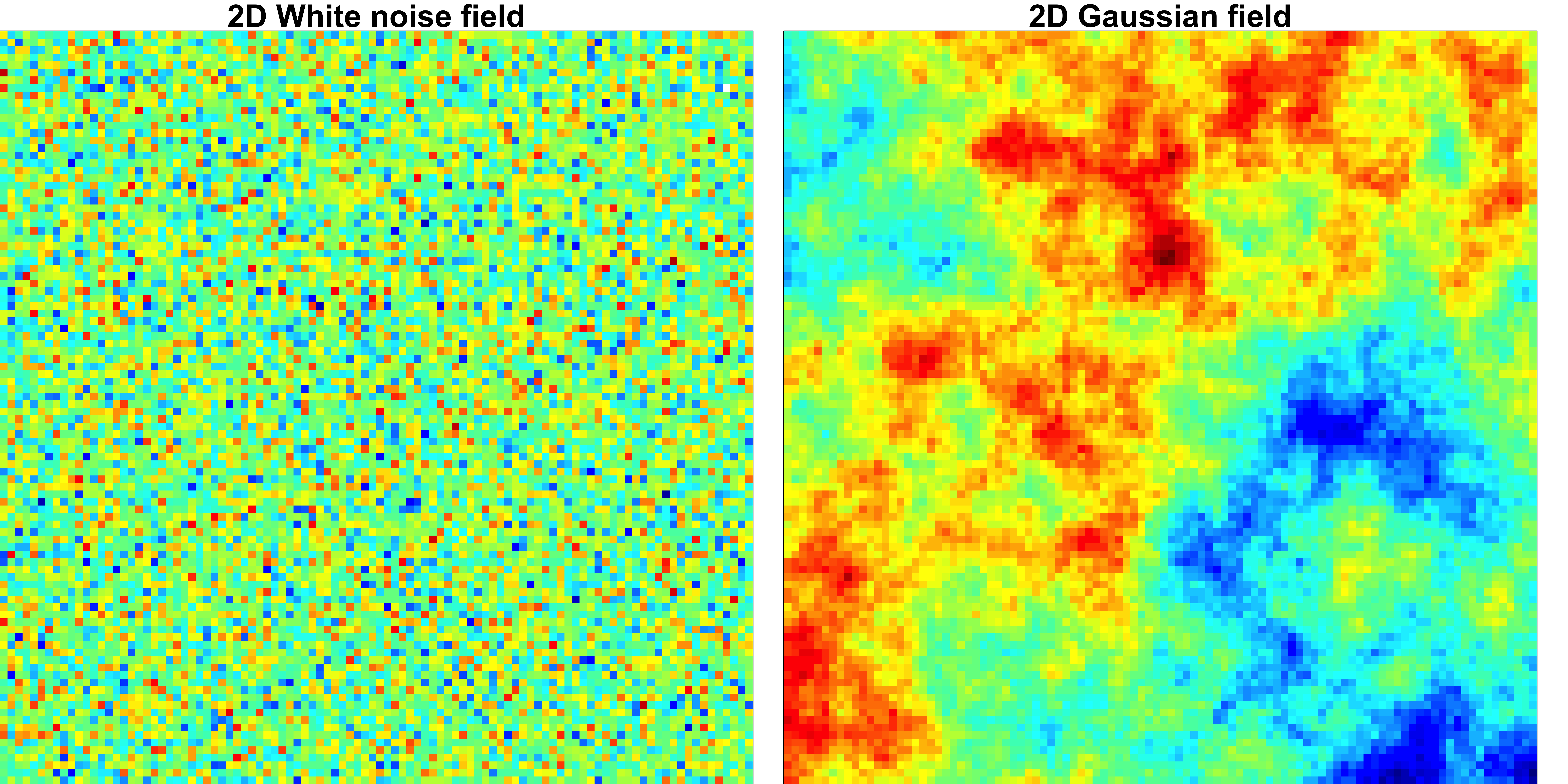
A stochastic process \(f(x), x \in \mathcal{X} \subset \mathbb{R}^D\), is a function whose values are random variables, for any value of \(x\).
Usually for \(D = 1\), the process is a temporal process, and for \(D > 1\), it is referred to as a spatial process.
- A Gaussian process (GP) is a process where all finite-dimensional distributions are multivariate Gaussian, for any choice of \(n\) and \(x_1\dots, x_n \in \mathbb{R}^D\):
\[f(x_1), \dots, f(x_n) \sim N_n\left(\boldsymbol \mu, \boldsymbol \Sigma\right)\]
- Write \(Y(\cdot) \sim GP\) to denote that the function \(Y\) is a GP.
Mean and Covariance Function
- To fully specify a Gaussian distribution we need the mean and covariance, \(Y \sim N(\mu, \Sigma)\)
- To fully specify a Gaussian process we need the mean and covariance function, \[f(\cdot) \sim GP\left(m(\cdot), k(\cdot, \cdot)\right)\] where
\[\text{E}(f(x)) = m(x)\] \[\text{Cov}(f(x), f(x')) = k(x, x')\]
Popular choices of \(m(\cdot)\) are \(m(x) = 0\) or \(m(x) = \text{const}\) for all \(x\), or \(m(x) = \beta'x\)
Care more about the covariance function or kernel function as it governs the how the process looks like by defining the similarity between data points.
Covariance Function Kernel
\[\text{Cov}(f(x), f(x')) = k(x, x')\]
-
\(k(x, x')\) must be a positive semi-definite function, leading to valid covariance matrices:
- Given locations \(x_1, \dots, x_n\), the \(n \times n\) Gram matrix \(K\) with \(K_{ij} = k(x_i, x_j)\) must be a positive semi-definite matrix.
- Often assume \(k(x, x')\) is a function of only the distance between locations: \[\text{Cov}(f(x), f(x')) = k(\|x-x'\|) = k(r)\]
- the GP is a stationary process.
- the covariance function is isotropic.
- The covariance function determines the nature of the GP, hypothesis space of functions.
Squared Exponential (SE) Kernel
\[k(x, x' \mid \tau, h) = \tau^2 \exp\left(-\frac{(x - x')^2}{2h^2} \right)\]
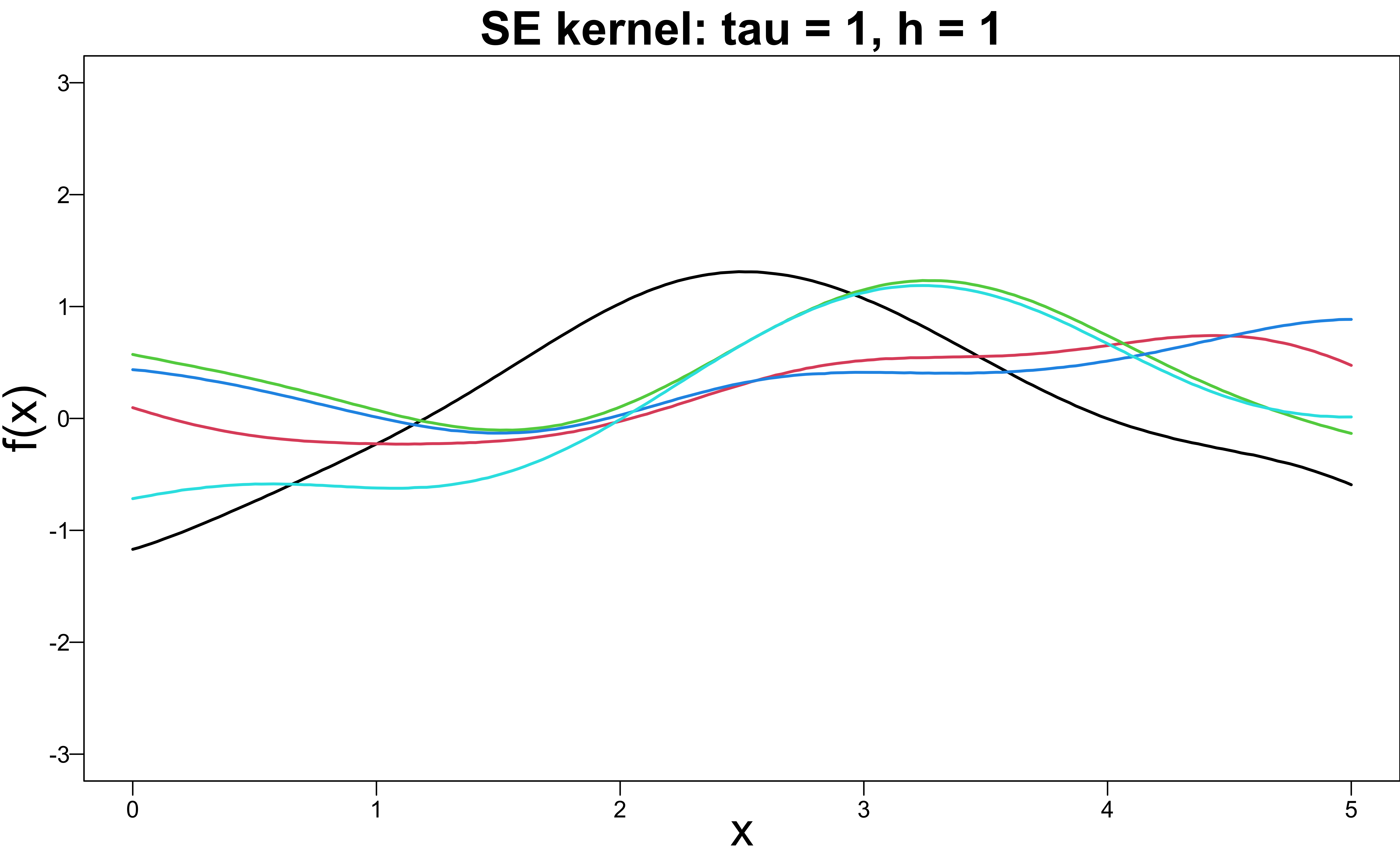
\[k(x, x') = \exp\left(-\frac{1}{2}(x - x')^2 \right)\]
Squared Exponential (SE) Kernel
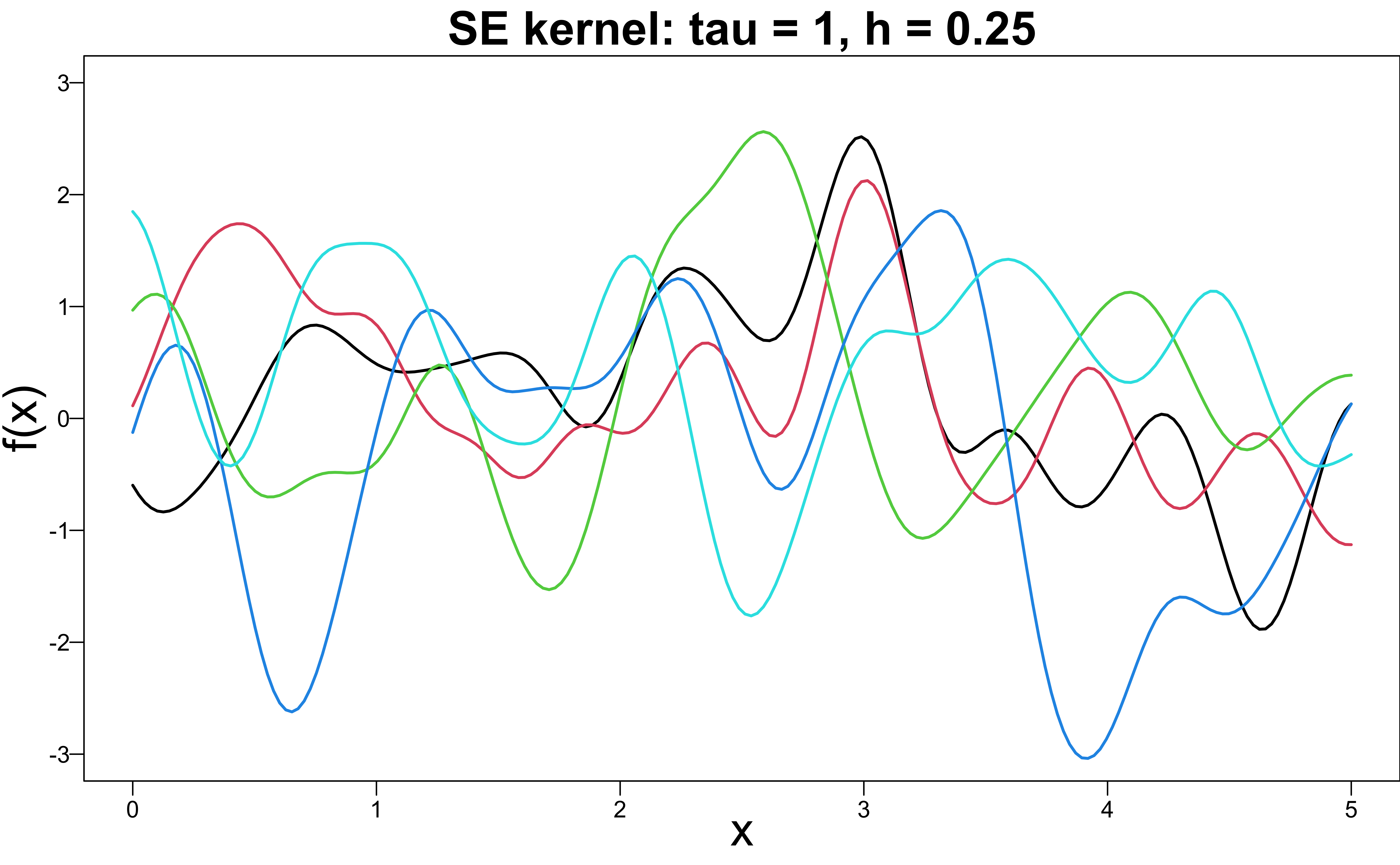
\[k(x, x') = \exp\left(-\frac{1}{2}\frac{(x - x')^2}{ {\color{red}{0.25}}^2} \right)\]
- The parameter \(h\) is the characteristic length-scale that controls the number of level-zero upcrossings.
Squared Exponential (SE) Kernel
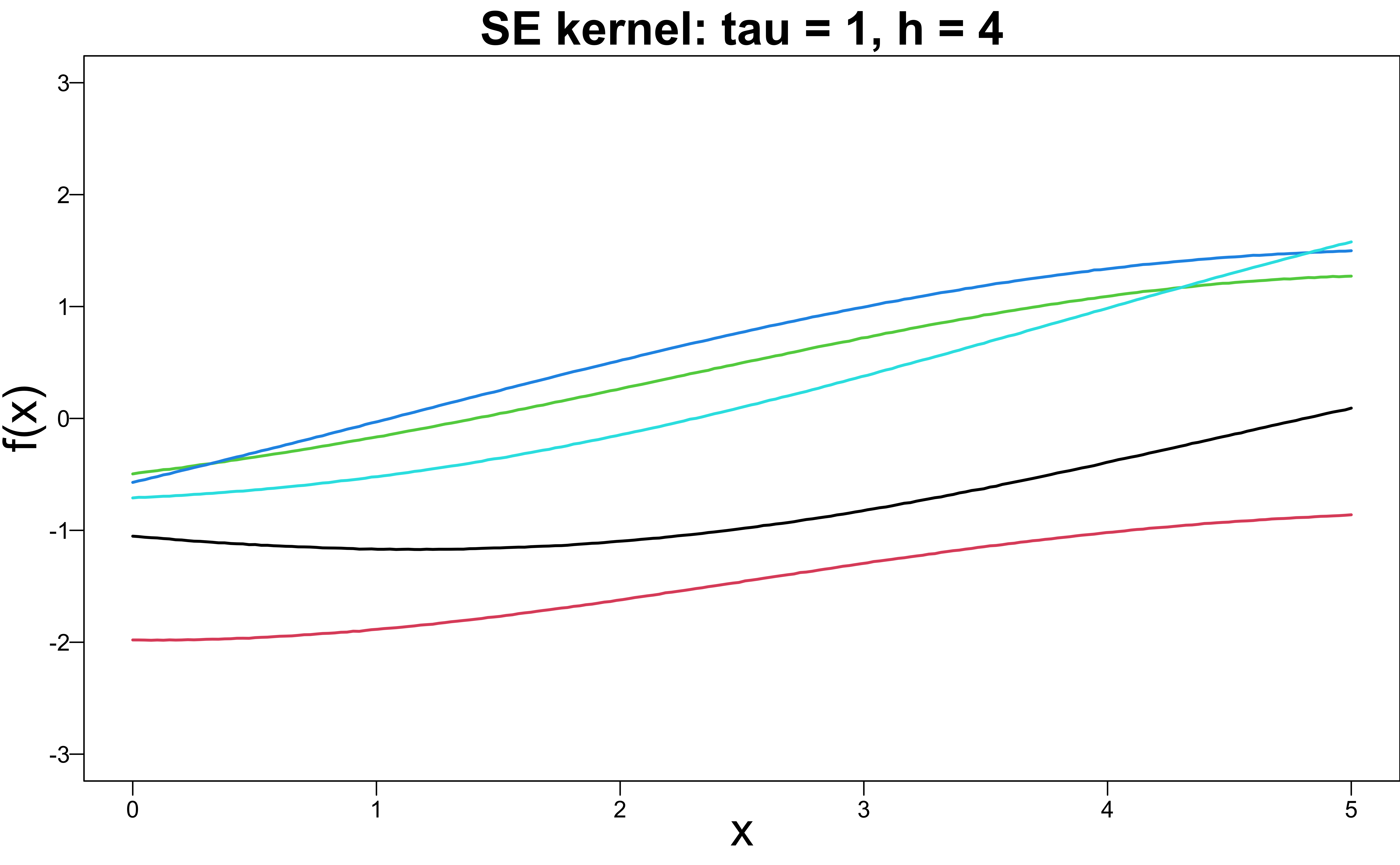
\[k(x, x') = \exp\left(-\frac{1}{2}\frac{(x - x')^2}{ {\color{red}{4}}^2} \right)\]
Squared Exponential (SE) Kernel
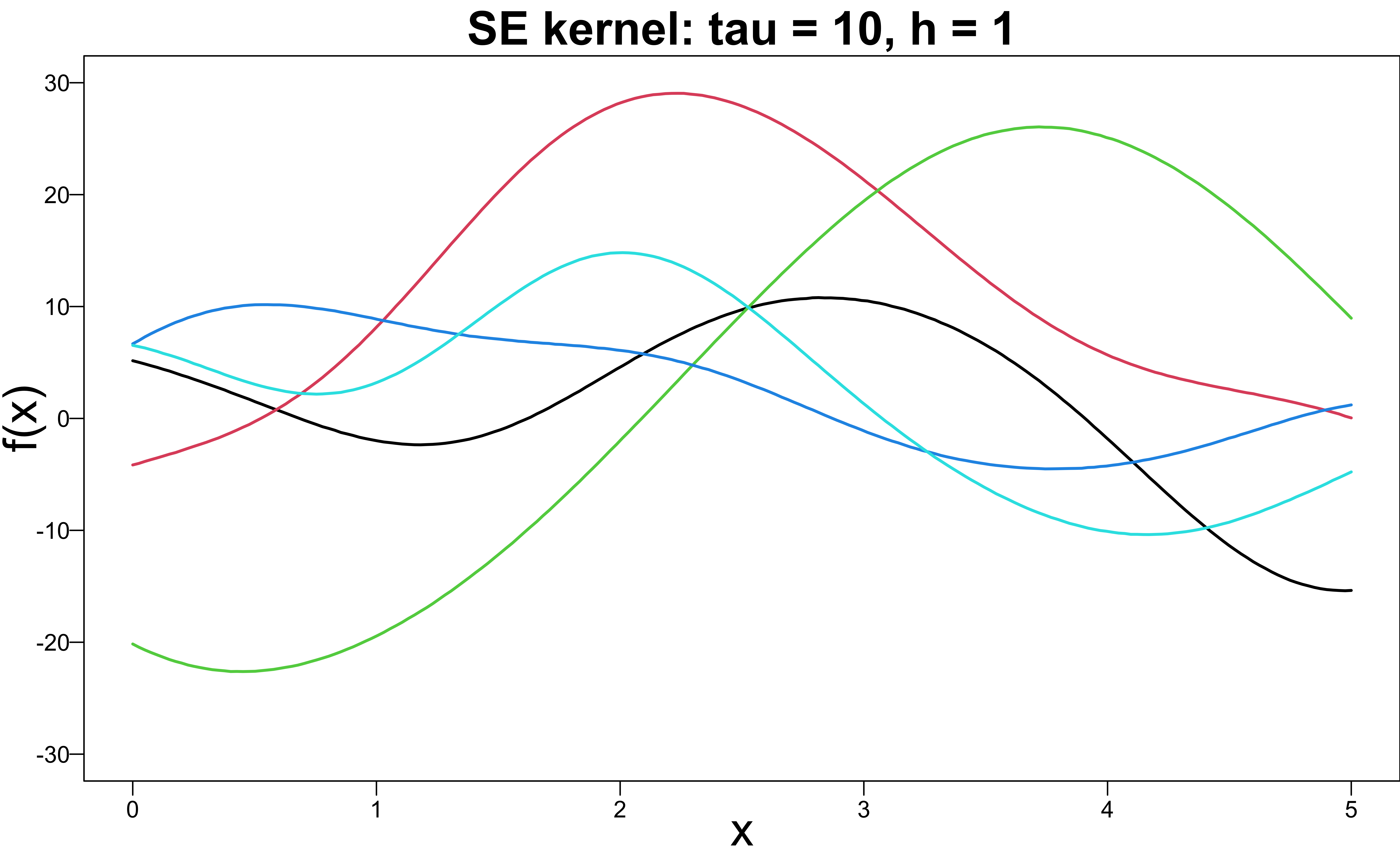
\[k(x, x') = {\color{red}{10^2}}\exp\left(-\frac{1}{2}(x - x')^2 \right)\]
- The parameter \(\tau\) is the variance of \(f(x)\) that controls the vertical variation of the process.
Exponential Kernel
\[k(x, x' \mid \tau, h) = \tau^2 \exp\left(-\frac{|x - x'|}{h} \right)\]
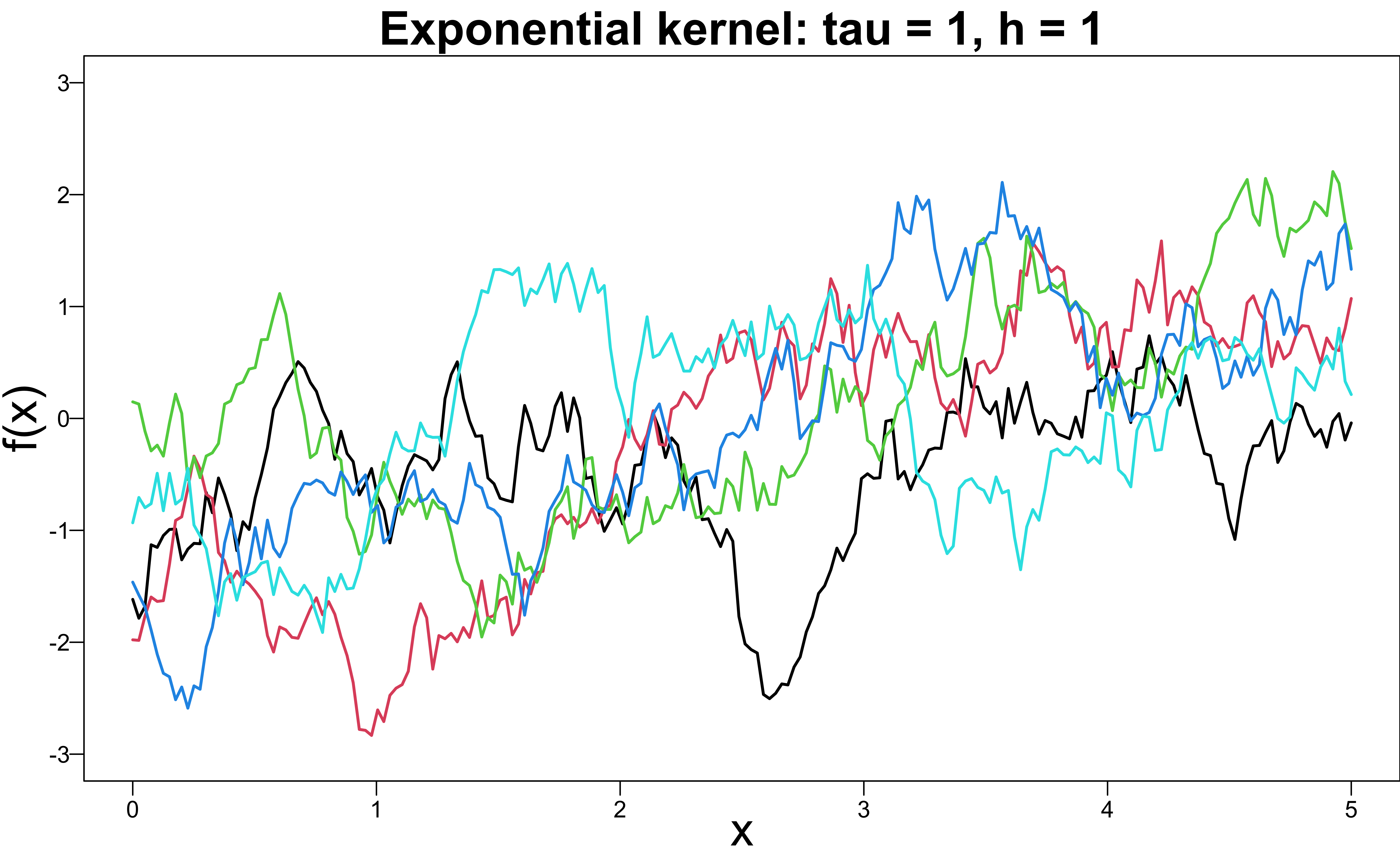
Brownian Motion
\[k(x, x') = \min(x, x')\]

White Noise
\[k(x, x') = \begin{cases} 1 & \quad \text{if } x = x' \\ 0 & \quad \text{otherwise} \end{cases}\]
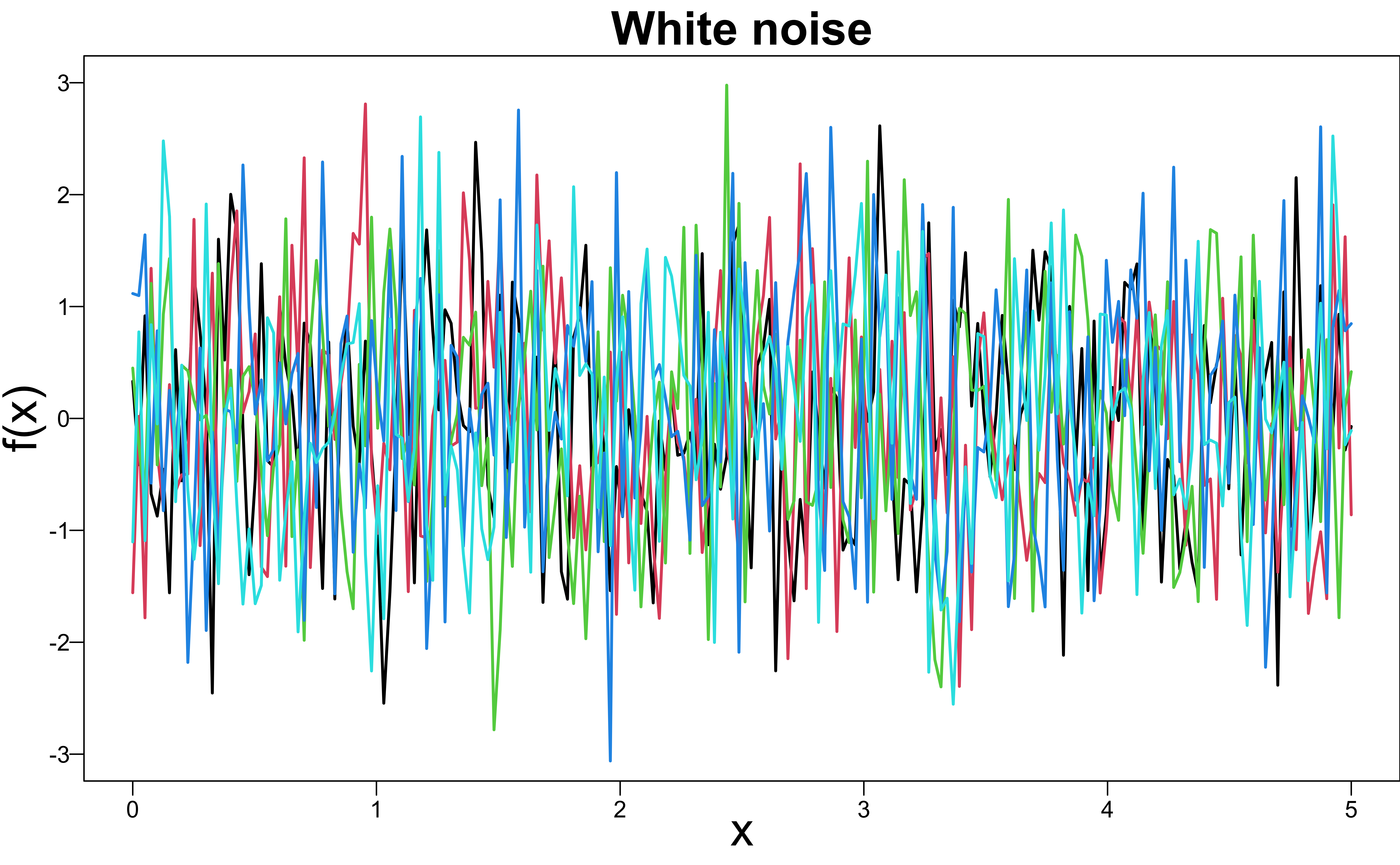
Why GP?
- The GP inherits its properties primarily from the covariance function \(k\):
- Smoothness
- Differentiability
- Variance
- Sums of Gaussians are Gaussian.
- Marginal distributions of multivariate Gaussians are still Gaussian.
- Any affine transformation of a Gaussian is a Gaussian.
- Conditional distributions are still Gaussian.
\[\mathbf{Y}= \begin{pmatrix} \mathbf{Y}_1 \\ \mathbf{Y}_2 \end{pmatrix} \sim N\left(\boldsymbol \mu, \boldsymbol \Sigma\right), \quad \boldsymbol \mu= \begin{pmatrix} \boldsymbol \mu_1 \\ \boldsymbol \mu_2 \end{pmatrix}, \quad \boldsymbol \Sigma= \begin{pmatrix} \Sigma_{11} & \Sigma_{12}\\ \Sigma_{21} & \Sigma_{22} \end{pmatrix}\] \[(\mathbf{Y}_2 \mid \mathbf{y}_1 = \mathbf{y}_1) \sim N\left(\boldsymbol \mu_2 + \Sigma_{21} \Sigma_{11}^{-1} (\mathbf{y}_1 - \boldsymbol \mu_1), \Sigma_{22} - \Sigma_{21}\Sigma_{11}^{-1}\Sigma_{12}\right)\]
Conditional Updates of Gaussian Processes
- If \(f(\cdot) \sim GP\),
\[f(x_1), \dots, f(x_n), f(x) \sim N(\boldsymbol \mu, \boldsymbol \Sigma)\]
- If we observe its value at \(x_1, \dots, x_n\), then
\[f(x) \mid f(x_1), \dots, f(x_n) \sim N(\boldsymbol \mu^*, \boldsymbol \Sigma^*)\] where \(\boldsymbol \mu^*\) and \(\boldsymbol \Sigma^*\) are as on the previous slide.
- We still believe \(f\) is a GP even we’ve observed its value at a number of locations.
Gaussian Process Regression
Bayesian Conditioning Updates of GP: Prior
- Instead of assigning priors to parameters in the regression function, we assign a function prior to the regression function:
\[f(\cdot) \sim GP(0, k(\cdot, \cdot))\]
- For any points \(x_1, \dots, x_n, x^*\),
\[f(x_1), \dots, f(x_n), f(x^*) \sim N\left(0, \boldsymbol \Sigma\right)\]
\[\boldsymbol \Sigma= \left(\begin{array}{ccc|c} k(x_1, x_1) & \cdots & k(x_1, x_n) & k(x_1, x^*) \\ \vdots & & \vdots & \vdots \\ k(x_n, x_1) & \cdots & k(x_n, x_n) & k(x_n, x^*) \\ \hline k(x^*, x_1) & \cdots & k(x^*, x_n) & k(x^*, x^*) \end{array} \right) = \left(\begin{array}{ccc|c} & & & \\ & K & & K_* \\ & & & \\ \hline & K_*^T & & K_{**} \end{array} \right) \]
Bayesian Conditioning Updates of GP: Posterior
- Given observed information \(f(x_1), \dots, f(x_n)\)
\[f(x^*) \mid f(x_1), \dots, f(x_n) \sim N\left( \boldsymbol \mu^*, \boldsymbol \Sigma^*\right)\] where
\[\boldsymbol \mu^* = K_{*}^TK^{-1}{\bf f}\] with \({\bf f} = \left(f(x_1), \dots, f(x_n)\right)^T\)
\[\boldsymbol \Sigma^* = K_{**} - K_{*}^TK^{-1}K_{*}\]
No Noise/Nugget - Interpolation
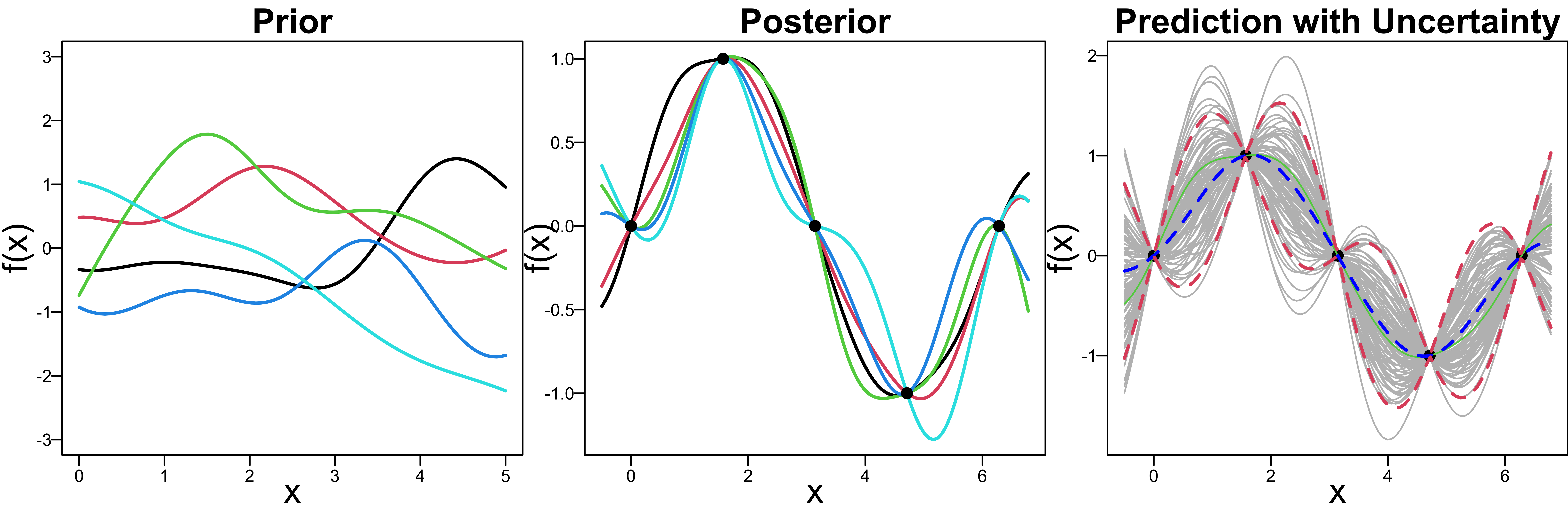
—–: posterior mean \(\boldsymbol \mu^*\)
—–: 95% posterior predictive interval \(\boldsymbol \mu^* \pm 1.96 \boldsymbol \Sigma^*\)
So far we treat \(f(x)\), the function values as data points.
There is no noise, and every posterior curve interpolates data points.
Noisy Observations/with Nugget - GP Regression (GPR)
- In reality, we don’t or can’t observe \(f(x)\) and like to estimate it.
\[\begin{align} y_i &= f(x_i) + \epsilon_i, \quad \epsilon_i\sim N(0, \sigma^2) \\ f(\cdot) &\sim GP(0, k(\cdot, \cdot; \theta))\end{align}\]
$$$$
\[y_1, \dots, y_n, f(x^*) \sim N\left(0, \boldsymbol \Sigma\right), \quad \boldsymbol \Sigma= \left(\begin{array}{ccc|c} & & & \\ & K + \sigma^2I & & K_* \\ & & & \\ \hline & K_*^T & & K_{**} \end{array} \right) \]
\[f(x^*) \mid y_1, \dots, y_n \sim N\left( \boldsymbol \mu^*, \boldsymbol \Sigma^*\right), \quad \boldsymbol \mu^* = K_{*}^T{\color{red}{(K + \sigma^2I)^{-1}}}{\bf y}, \quad \boldsymbol \Sigma^* = K_{**} - K_{*}^T{\color{red}{(K + \sigma^2I)^{-1}}}K_{*}\] with \({\bf y} = \left(y_1, \dots, y_n\right)^T\)
Noisy Observations/with Nugget - GP Regression (GPR)
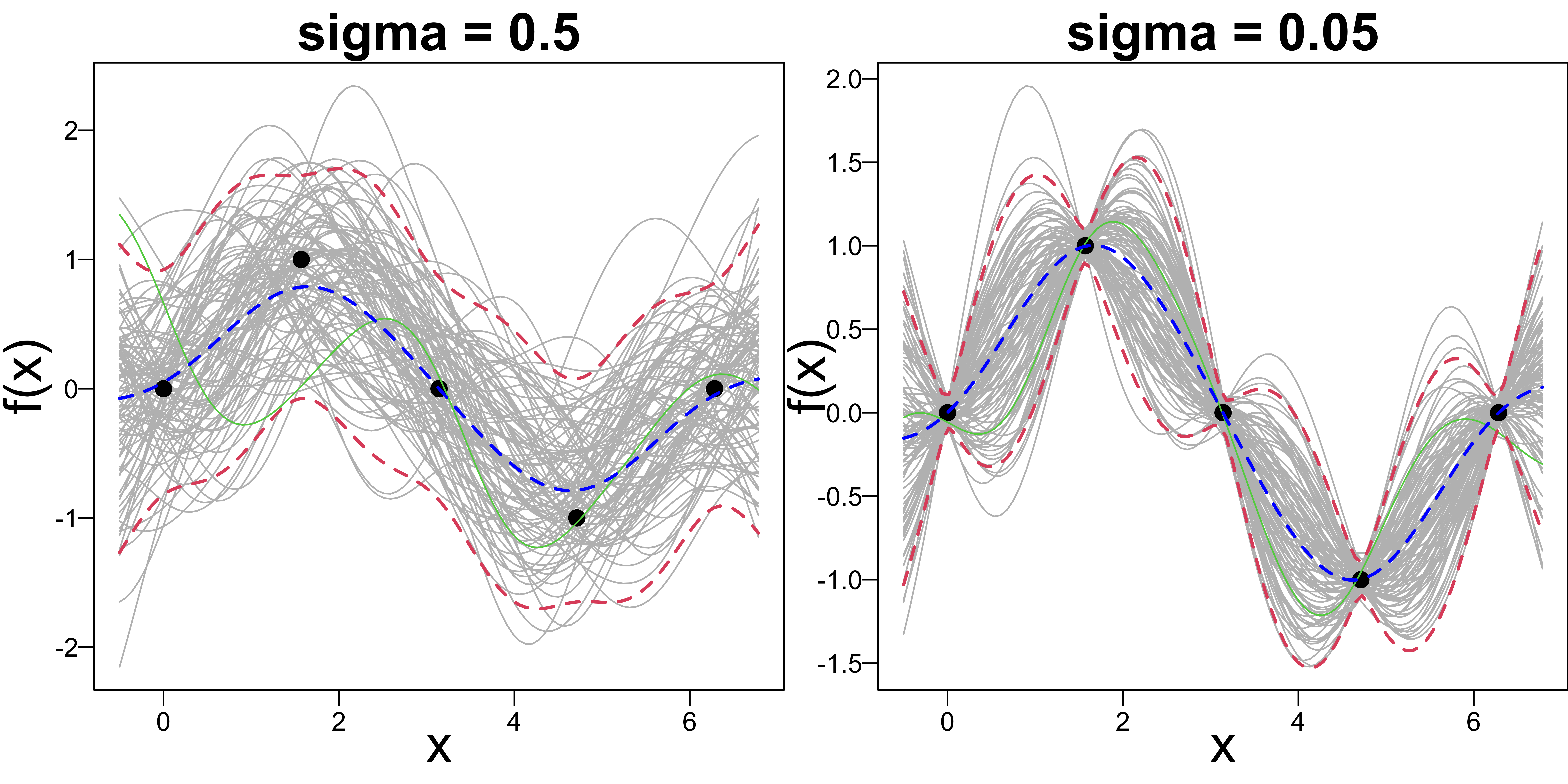
Hyperparameter Tuning

Empirical Bayes
Uses the observed data to estimate parameters \(\theta = (\sigma^2, \tau^2, h)\)
Find an empirical Bayes estimate for \(\theta\) from the marginal likelihood
\[ p\left( \mathbf{y}\mid \theta \right) = \int p\left( \mathbf{y}\mid {\bf f} \right) p\left( {\bf f} \mid \theta \right) \, d {\bf f} = N\left(\mathbf{0}, K(\tau, h)+\sigma^2I \right)\]
- \(\hat{\theta}_{EB} = \mathop{\mathrm{arg\,max}}_{\theta}\log p\left( \mathbf{y}\mid \theta \right)\).
Full Bayesian Inference
\[\begin{align} y_i &= f(x_i) + \epsilon_i, \, \, \epsilon_i \stackrel{\rm iid}{\sim} N(0, \sigma^2), \quad i = 1, \dots, n,\\ f(\cdot) &\sim GP\left(\mu, k(\cdot, \cdot)\right),\,\, \text{Cov}(f(x_i), f(x_j)) = k(x_i, x_j)\\ \sigma^2 &\sim IG(a_{\sigma}, b_{\sigma})\\ \tau^2 &\sim IG(a_{\tau}, b_{\tau})\\ h &\sim Ga(a_{h}, b_{h})\\ \mu & \sim N(0, b_{\mu}) \end{align}\]
- The model is Gibbsable, or the Metropolis-Hastings algorithm can be used when \({\bf f}\) is integrated out.