Dimension Reduction
MSSC 6250 Statistical Machine Learning
Department of Mathematical and Statistical Sciences
Marquette University
Unsupervised Learning
Unsupervised Learning
Supervised Learning: response \(Y\) and features \(X_1, X_2, \dots, X_p\) measured on \(n\) observations.
-
Unsupervised Learning: only features \(X_1, X_2, \dots, X_p\) measured on \(n\) observations.
- Not interested in prediction (no response to be predicted)
- Discover any interesting pattern or relationships among these features.
- Estimate the density, covariance, graph (network), etc. of \(\mathbf{X}\)
-
Dimension reduction for effective data visualization or extracting most important information those features contain.
- plot a bunch of points of \(\boldsymbol{x} = (x_1, x_2, \dots, x_p)\) in a 2-D scatter plot (manifold). (reduce dimension from \(p\) to 2)
- use 2 variables to explain most variations or represents high data density in the \(p\) variables
-
Clustering discovers unknown subgroups/clusters in data
- find 3 sub-groups of people based on variables income, occupation, age, etc
Background: Dimensions
One-Dimension (1D) Number line
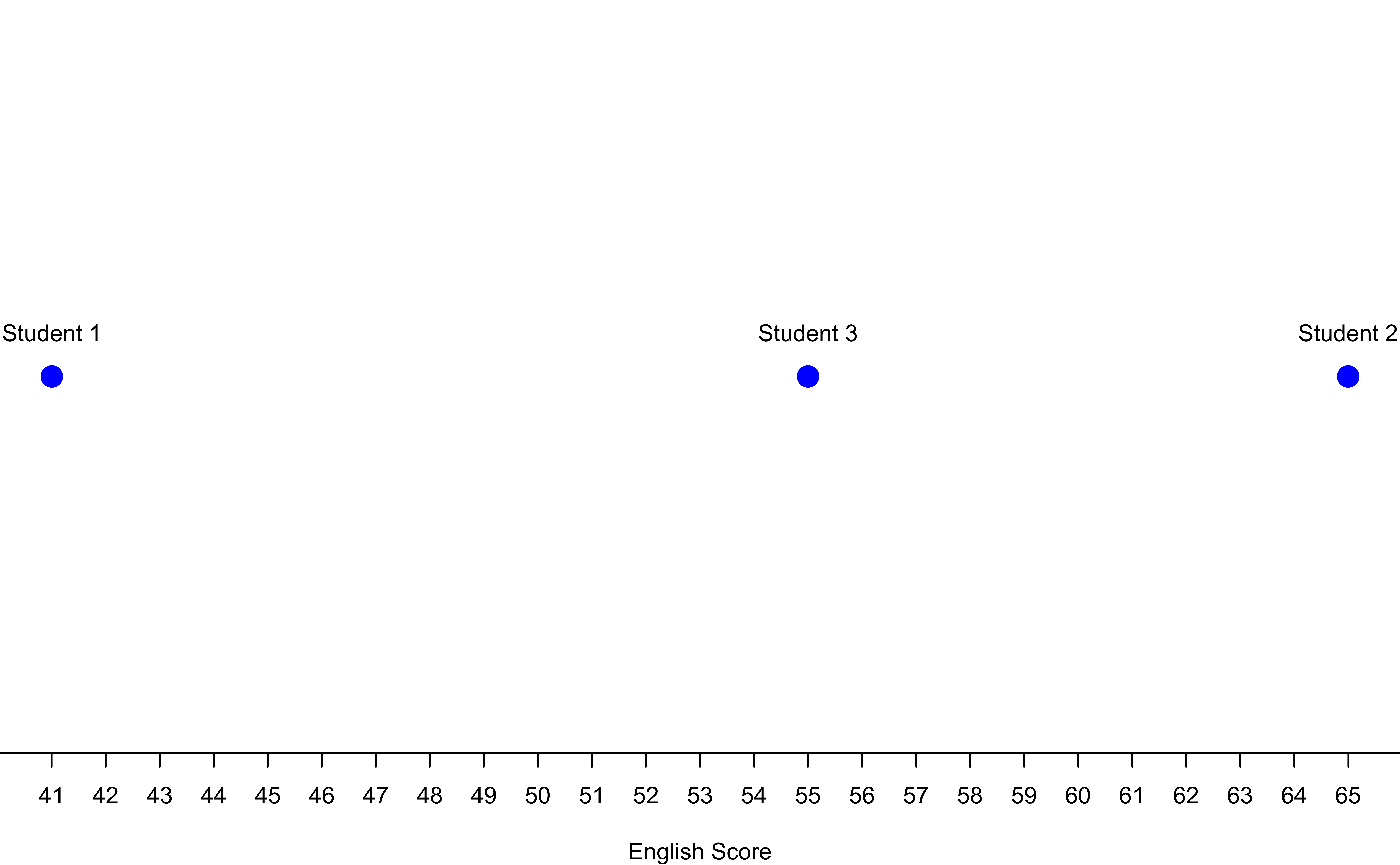
One-Dimension (1D) Number line: Uniform students
1D Number line: Non-uniform students
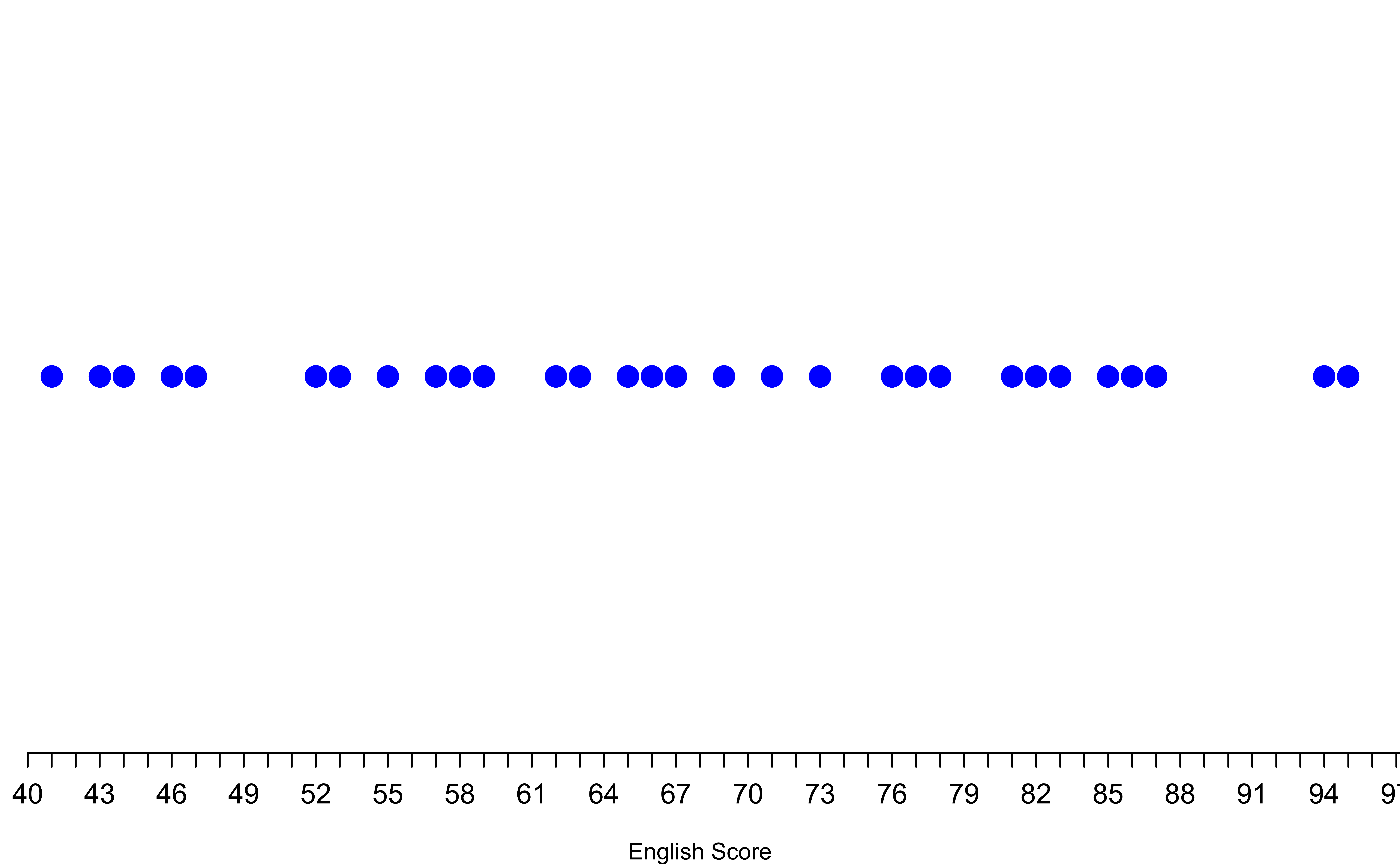
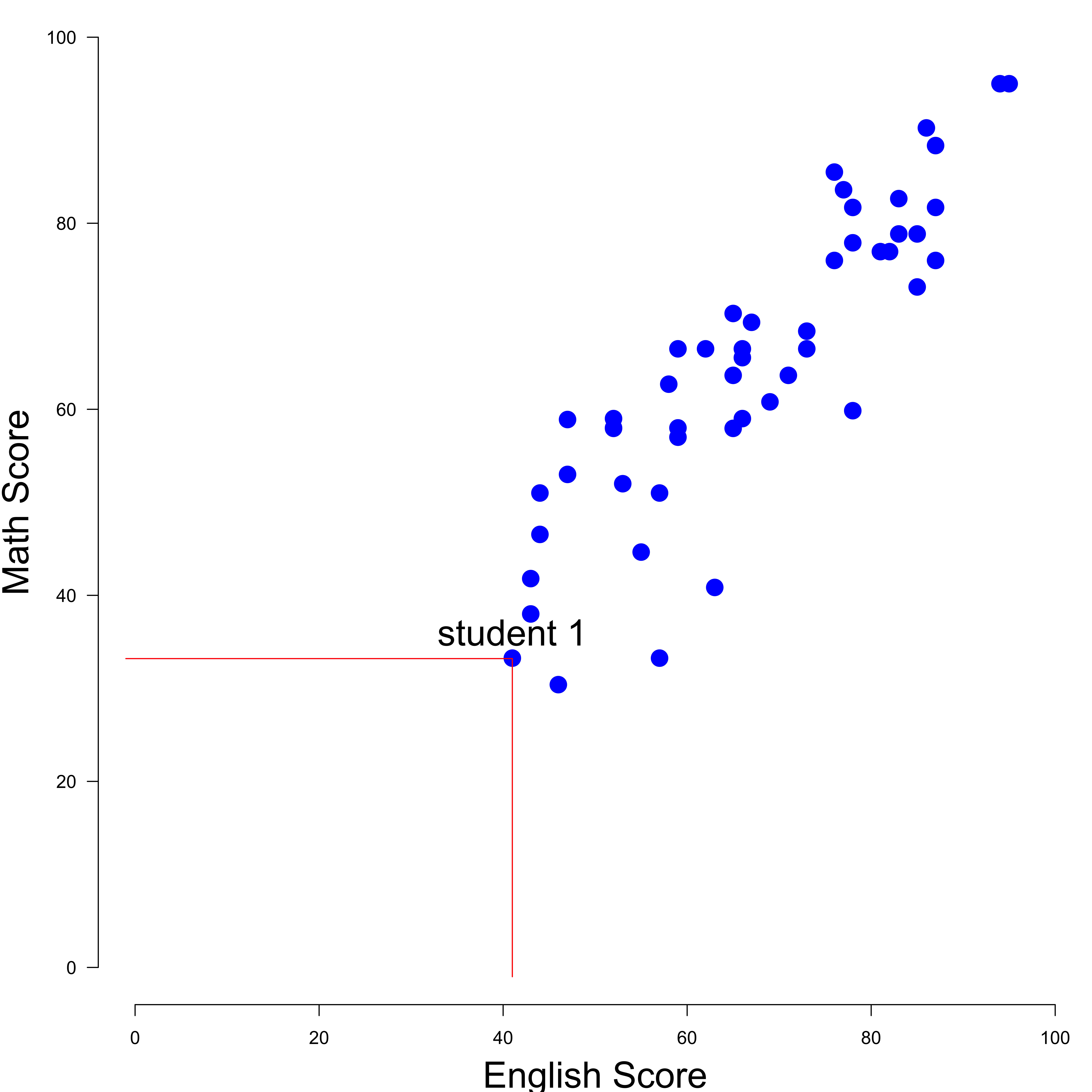
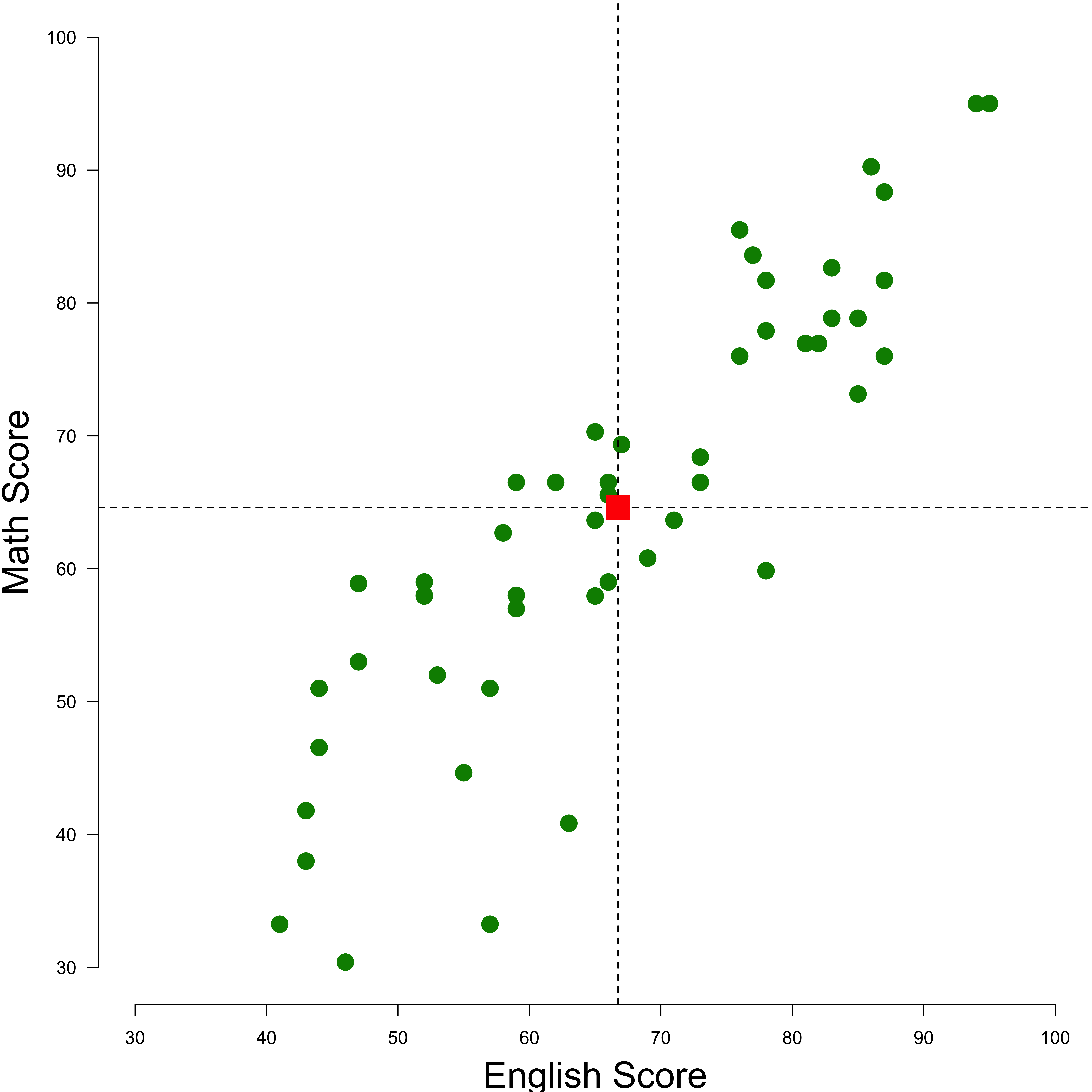
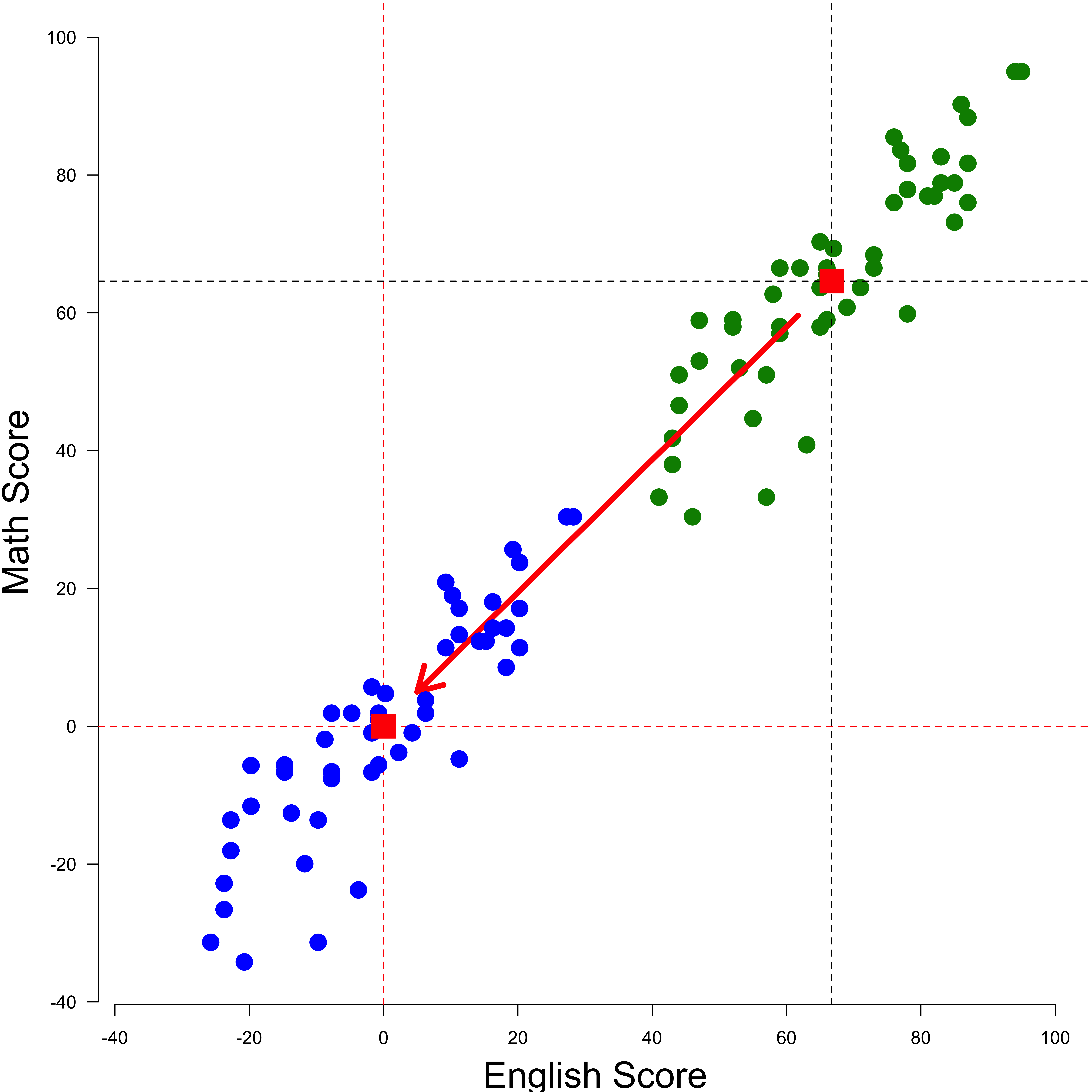
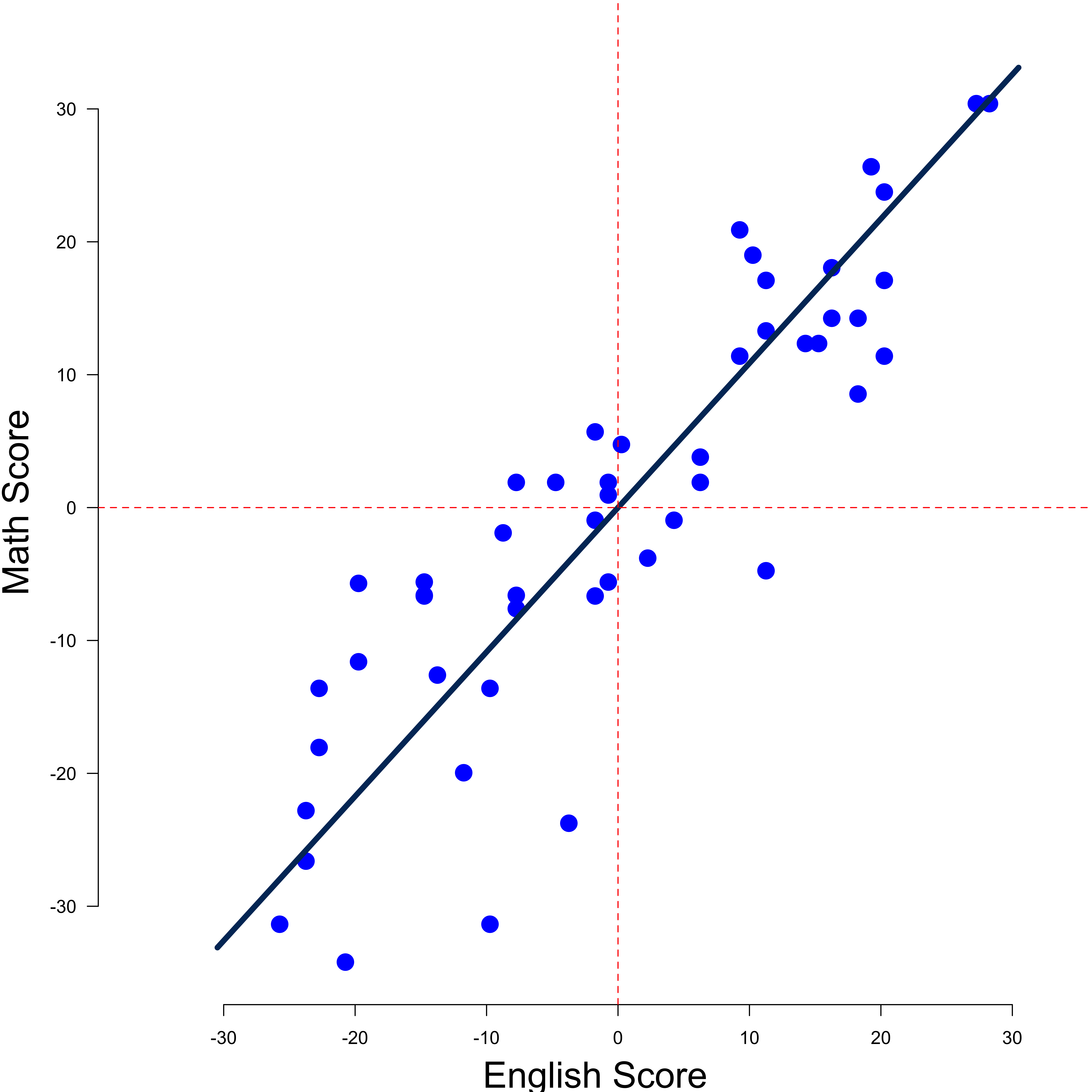
Two-Dimensions (2D) X-Y Scatter plot: High Correlated
English and Math measure an overall academic performance.
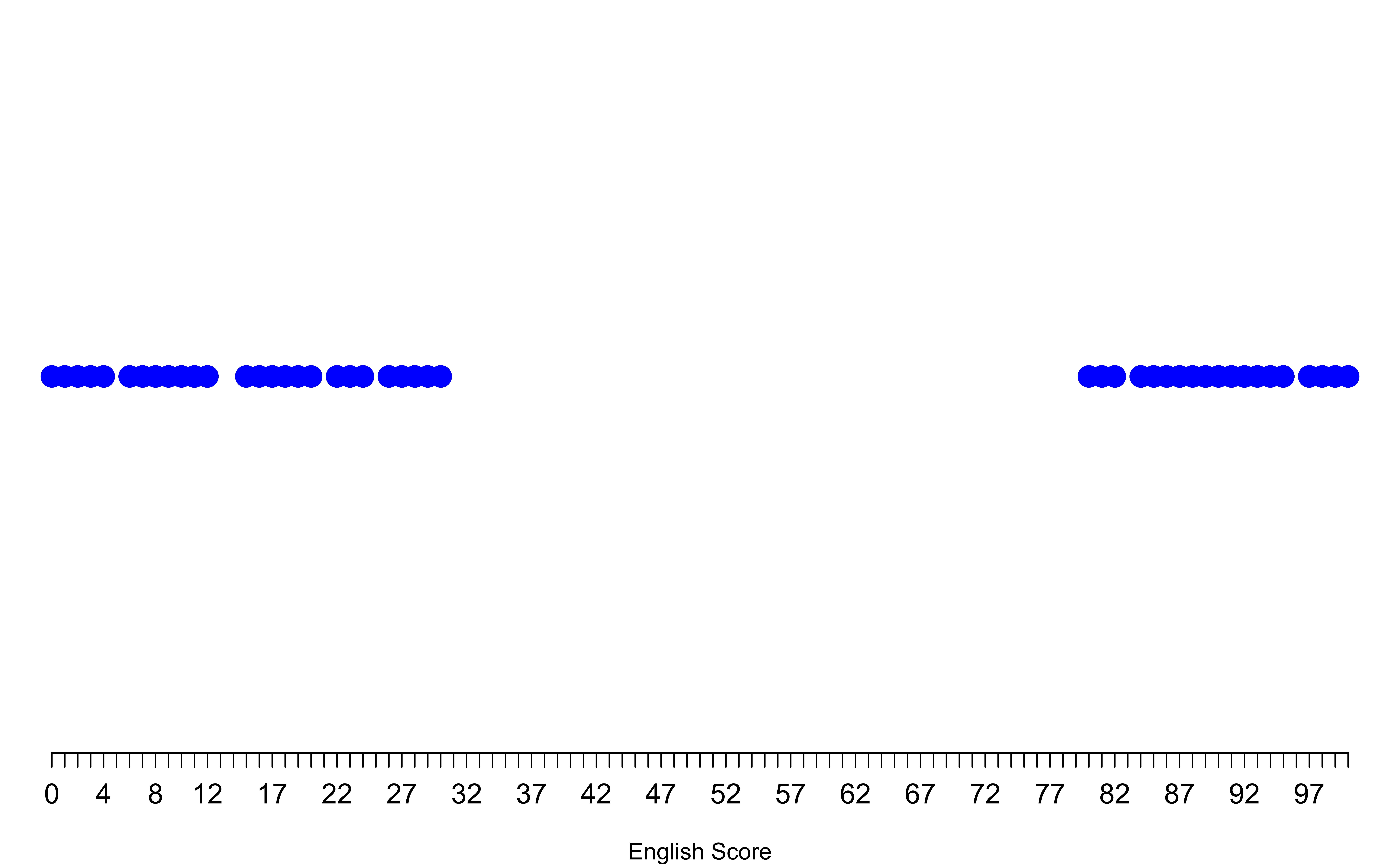
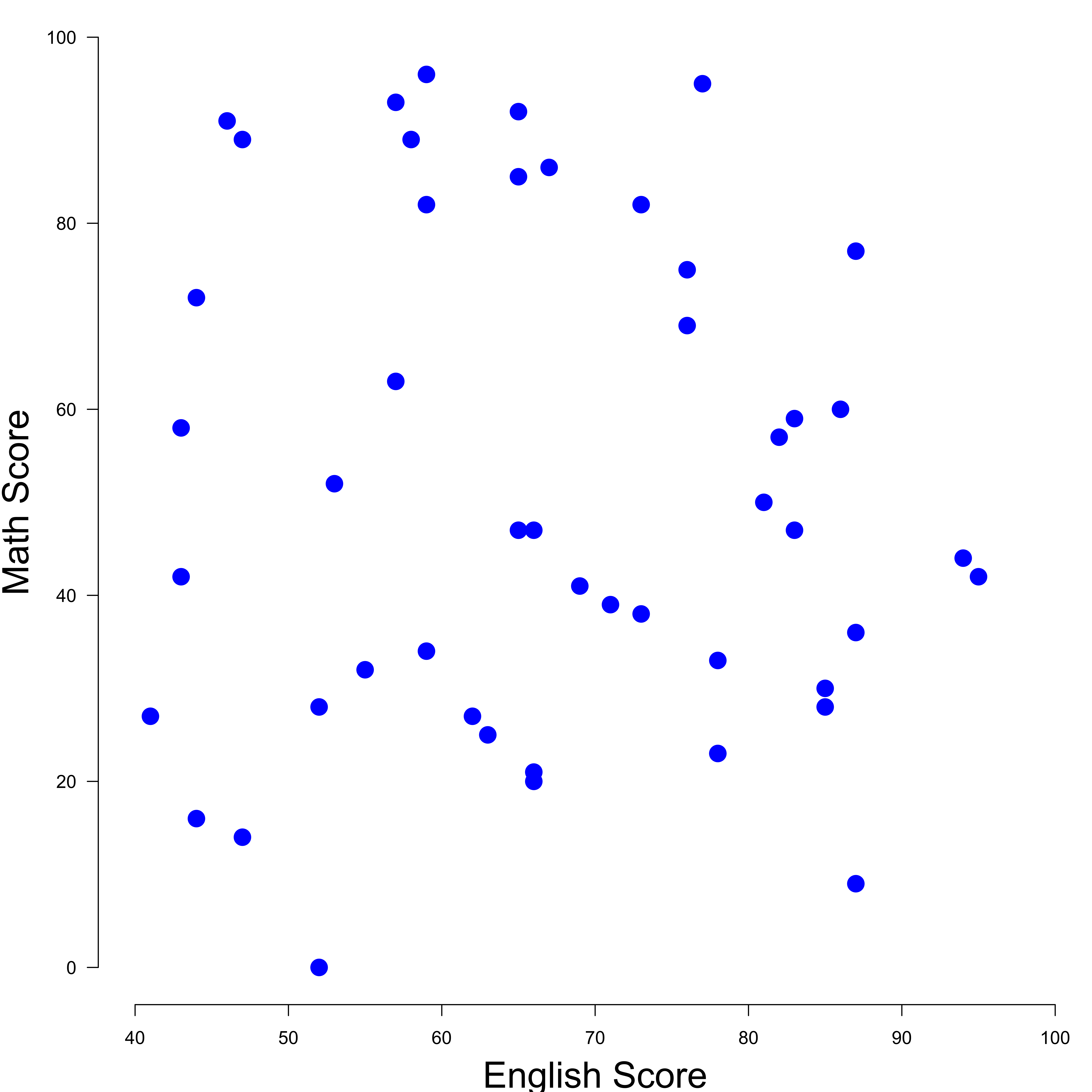
Two-Dimensions (2D) X-Y Scatter plot: No correlated
English and Math measure different abilities.
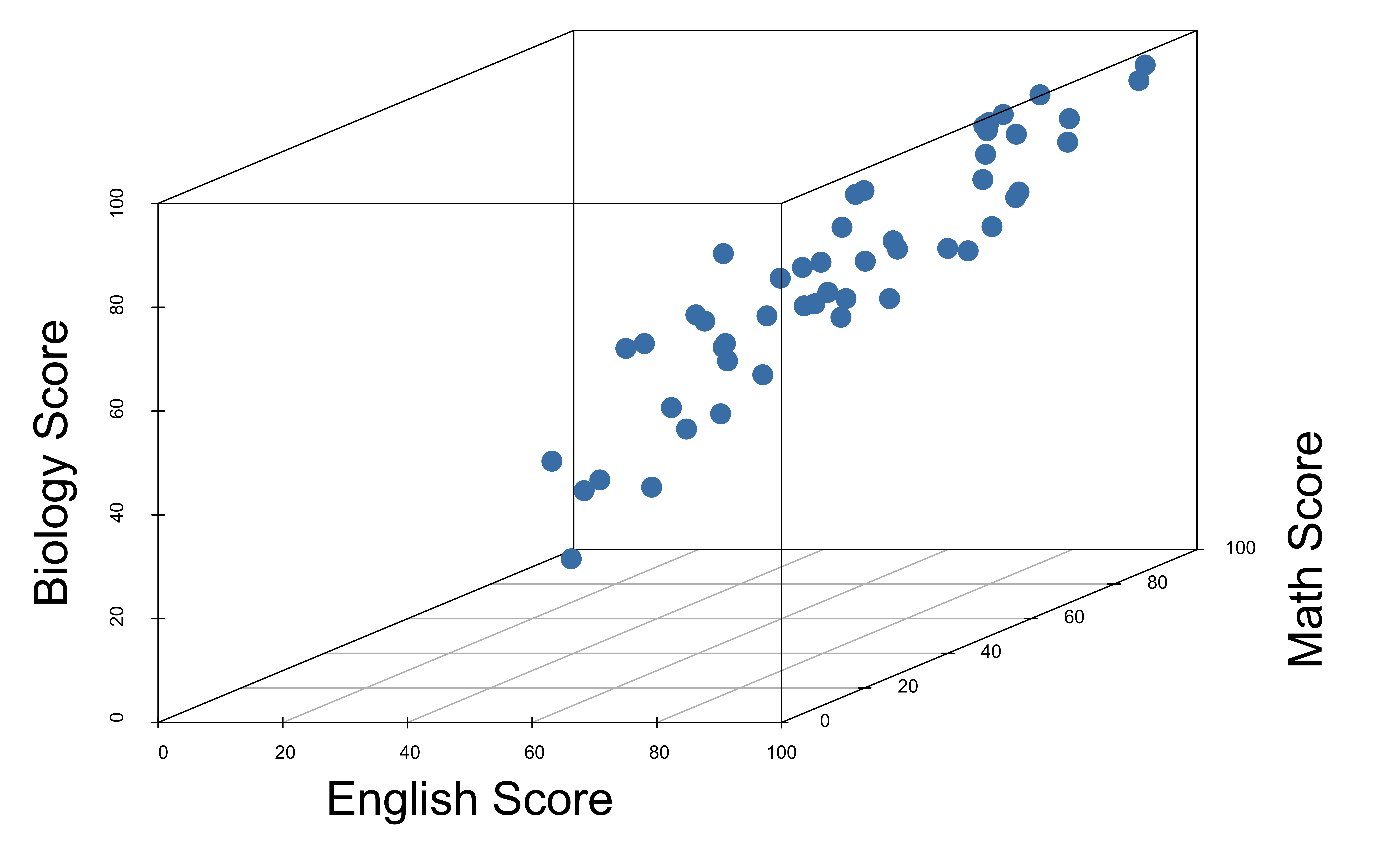
Three-Dimensions (3D) X-Y-Z Scatter plot
Four-Dimension (4D) X-Y-Z-? Scatter plot
How about Pair Plots?
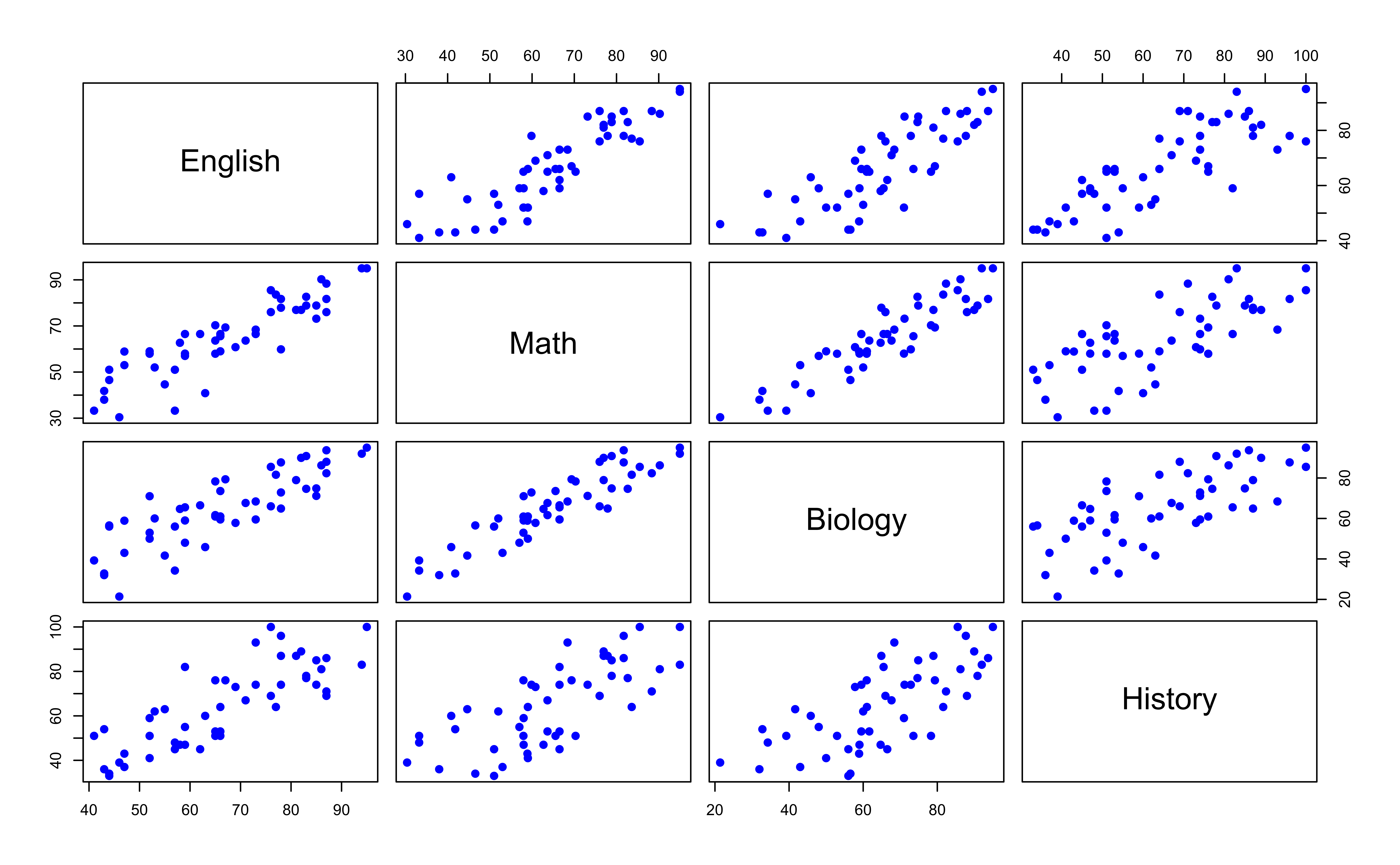
Tooooo Many Pair Plots!
- If we have \(p\) variables, there are \({p \choose 2} = p(p-1)/2\) pairs.
- If \(p = 10\), we have 45 such scatter plots to look at!
- In practice, we may encounter over 100 variables!!
Dimension Reduction
One variable represents one dimension.
With many variables in the data, we live in a high dimensional world.
GOAL:
Find a low-dimensional (usually 2D) representation of the data that captures as much of the information all of those variables provide as possible.
Use two created variables to represent all \(p\) variables, and make a scatter plot of the two created variables to learn what our observations look like as if they lived in the high dimensional space.
Why and when can we omit dimensions?
Variation mostly from One Variable
- Almost all of the variation in the data is from left to right.
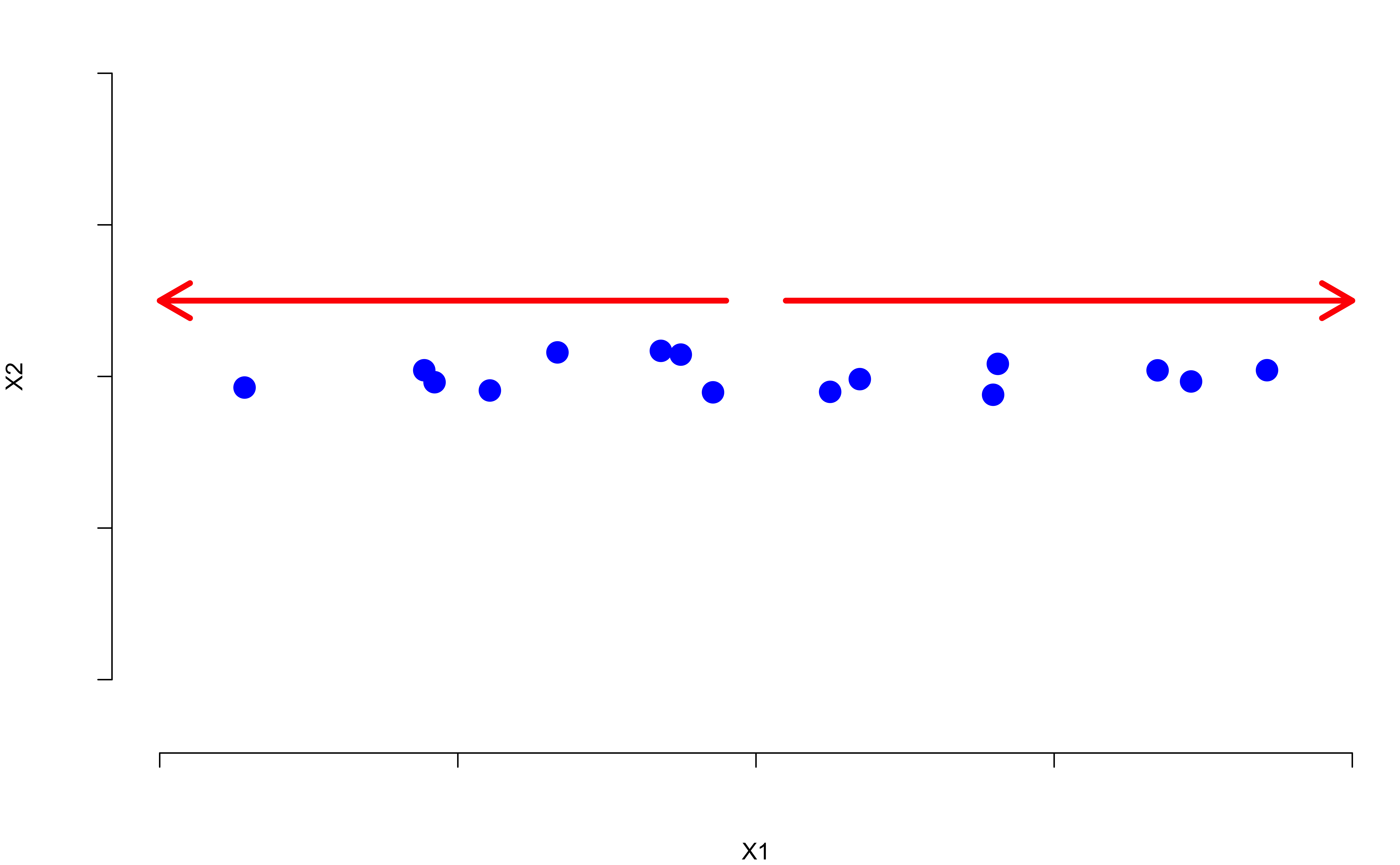
Variation mostly from One Variable
- If we flattened the data, the graph would not look much different.
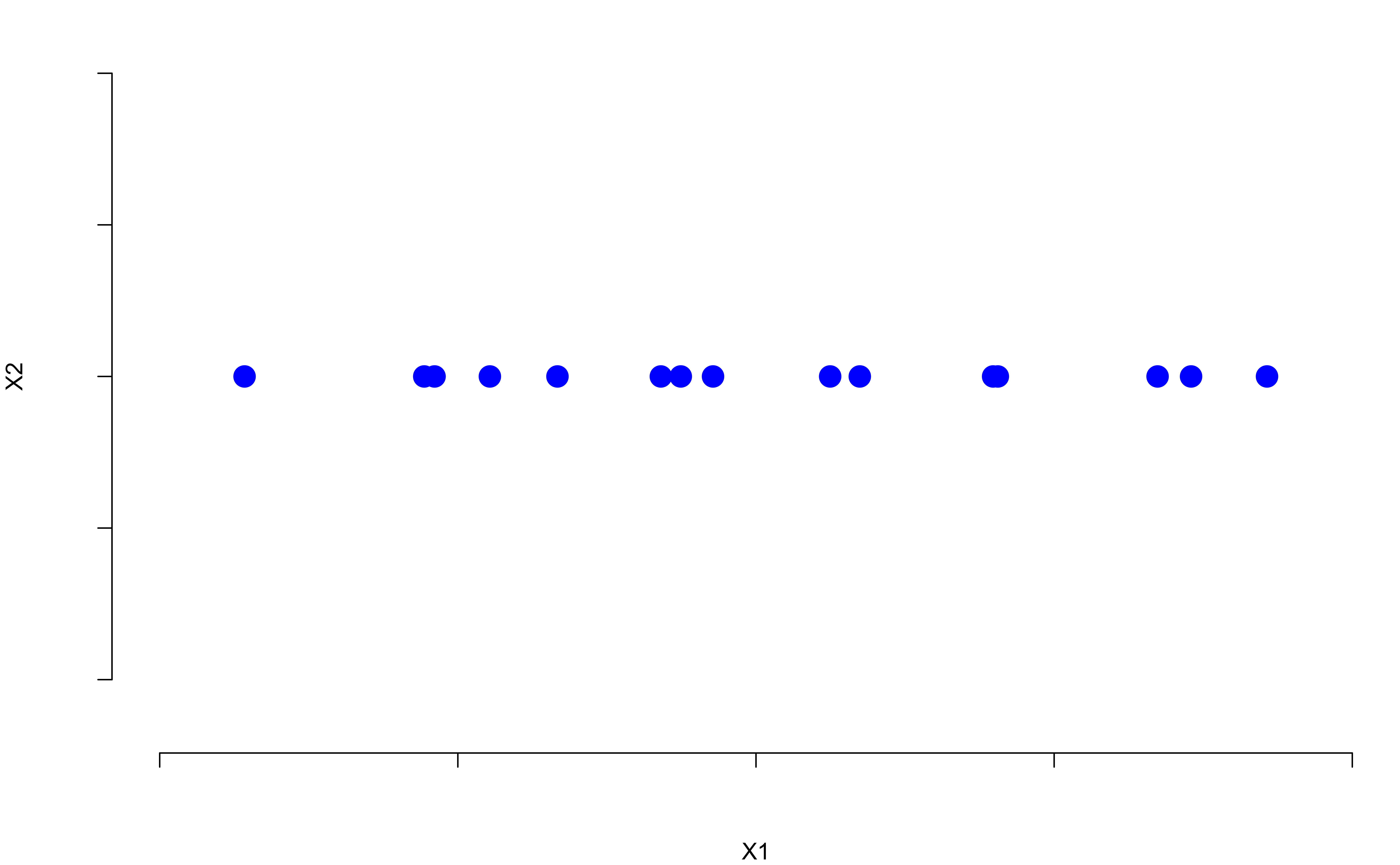
Variation mostly from One Variable
- If we flattened the data, we could graph it with a 1D number line!
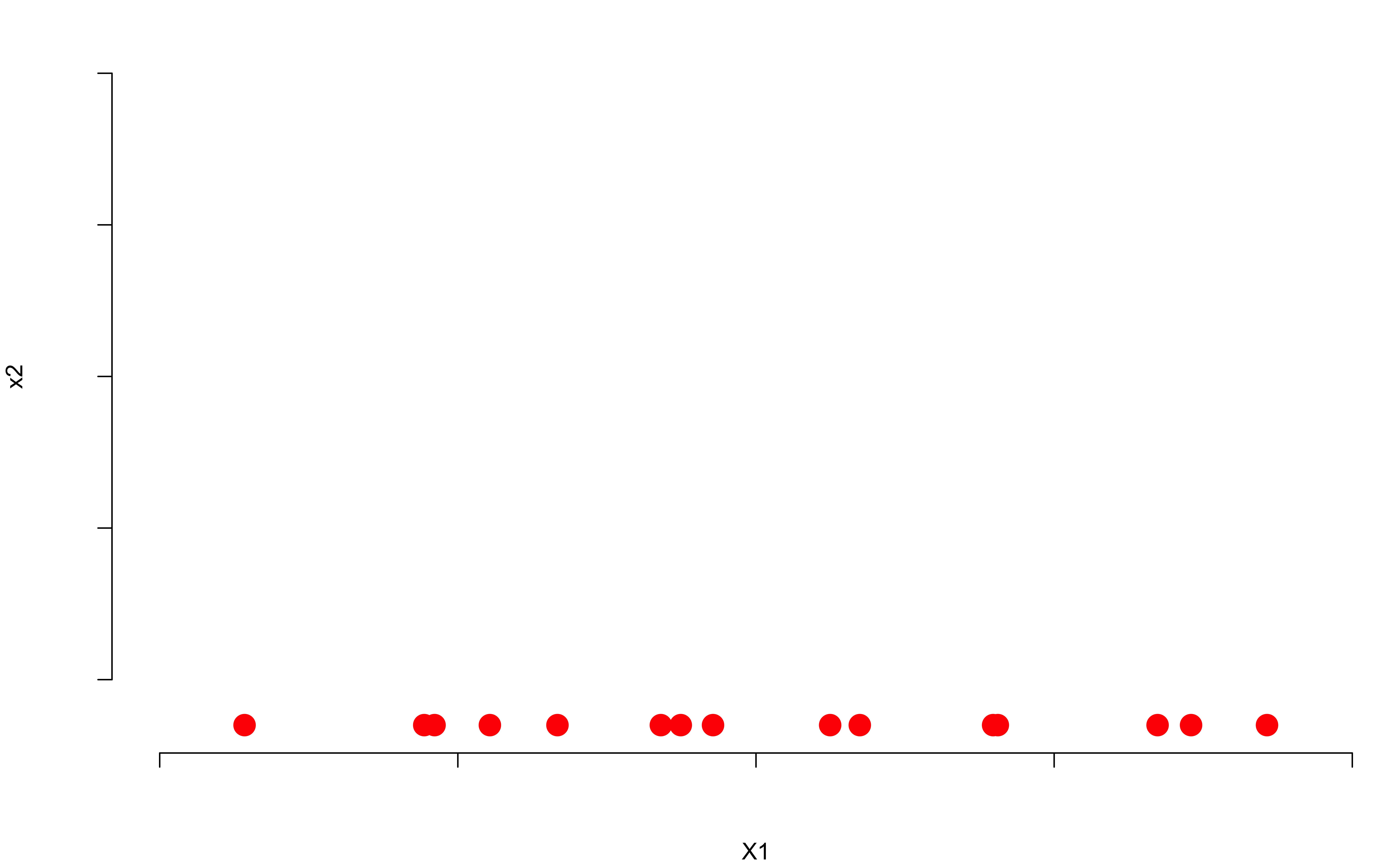
Variation mostly from One Variable
- Both graphs say “the important variation is left to right.”
Principal Component Analysis (PCA)
Idea of PCA
PCA is a dimension reduction tool that finds a low-dimensional representation of a data set that contains as much as possible of variation.
Each observation lives in a high-dimensional space (lots of variables), but not all of these dimensions (variables) are equally interesting/important.
The concept of interesting/important is measured by the amount that the observations vary along each dimension.
PCA Illustration: 2 Variable Example
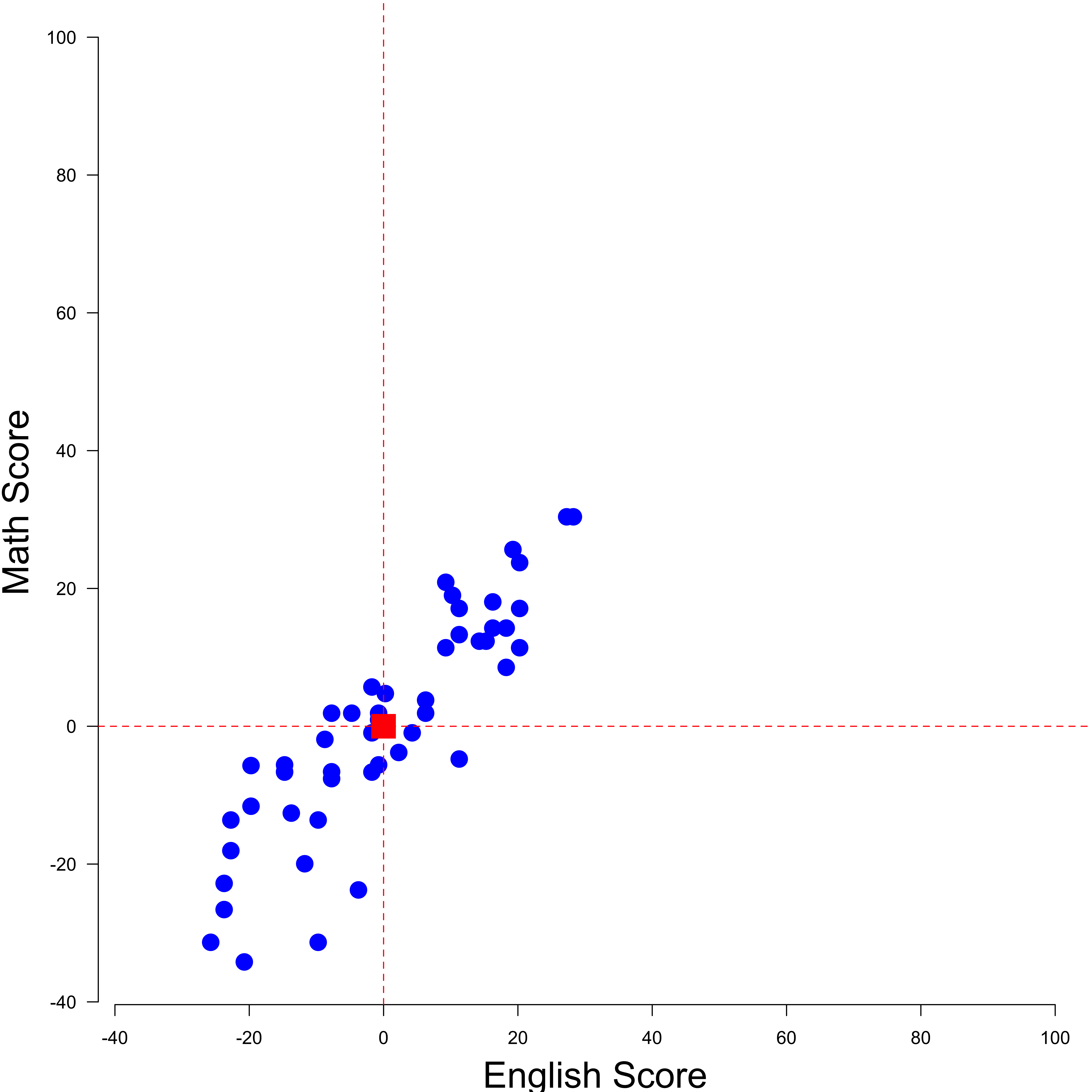
Step 1: Shift (or standardize) the Data
- So the two variables have both mean 0. If the variables are measured in a different unit, consider standardization, \(\frac{x_i - \bar{x}}{s_x}\).
- Shifting does not change how the data points are positioned relative to each other.
Step 2: Find a Line that Fits the Data the Best
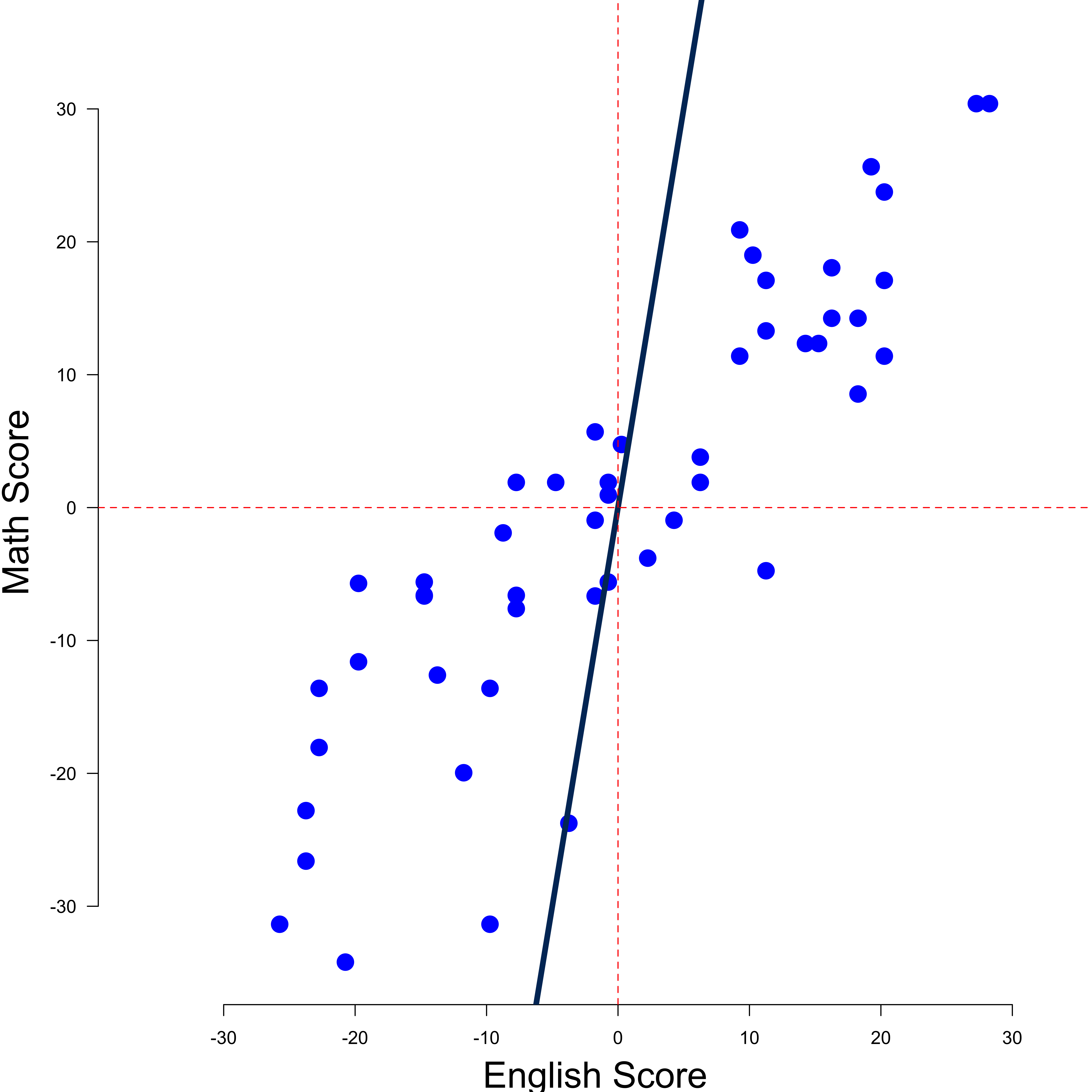
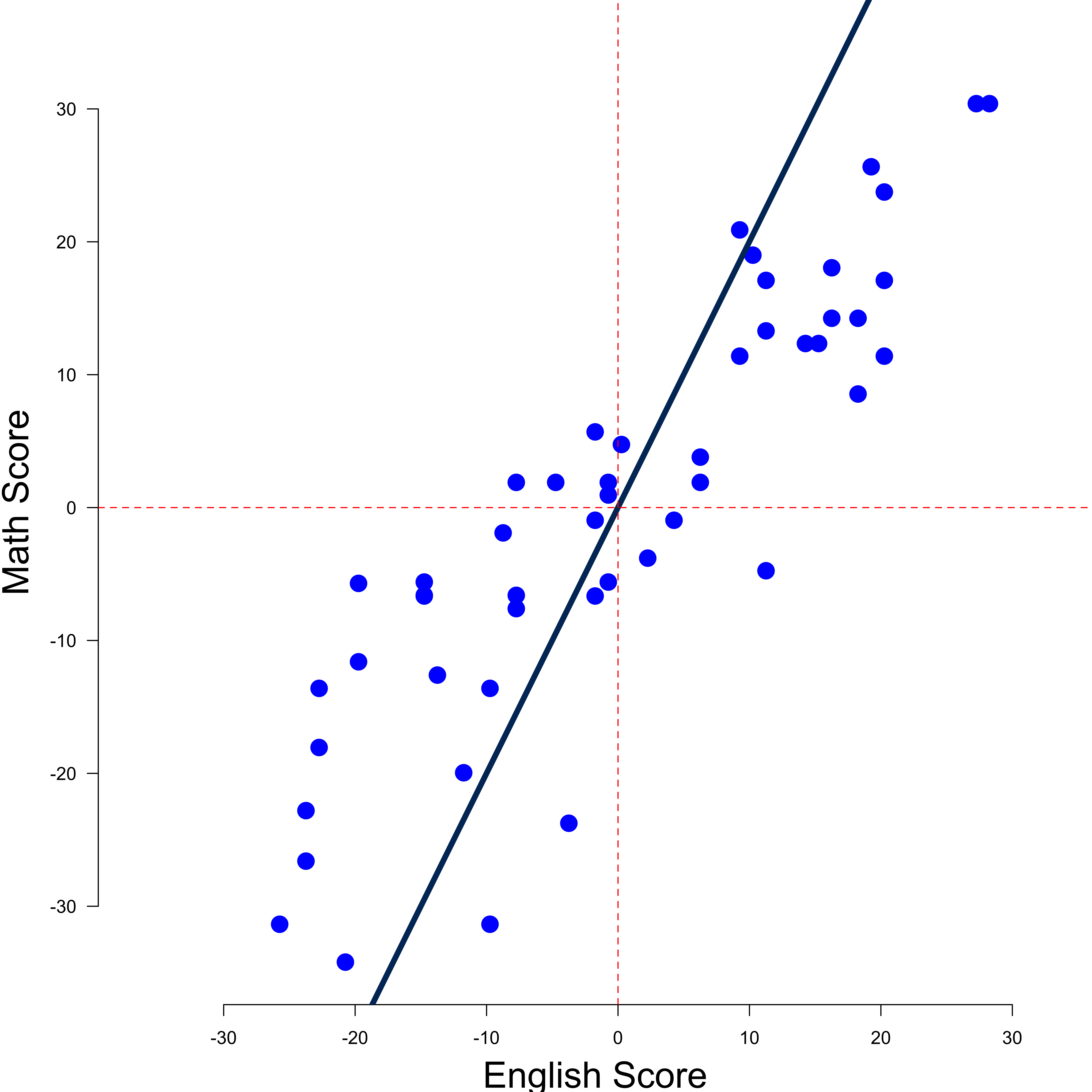
- Start with a line going through the origin.
- Rotate the line until it fits the data as well as it can, given that it goes through the origin.
Step 2: Find a Line that Fits the Data the Best
- Start with a line going through the origin.
- Rotate the line until it fits the data as well as it can, given that it goes through the origin.
Step 2: Find a Line that Fits the Data the Best
- Start with a line going through the origin.
- Rotate the line until it fits the data as well as it can, given that it goes through the origin.
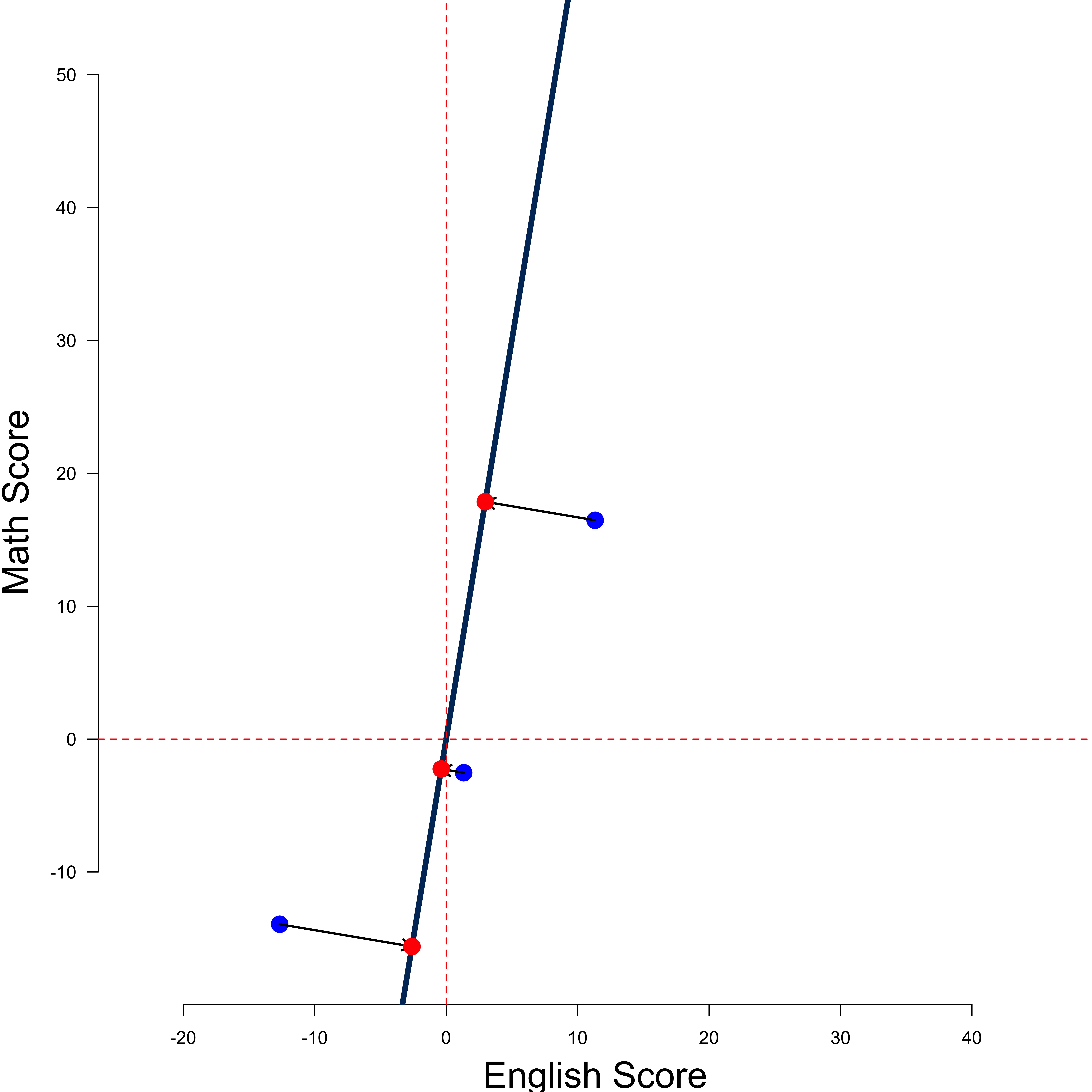
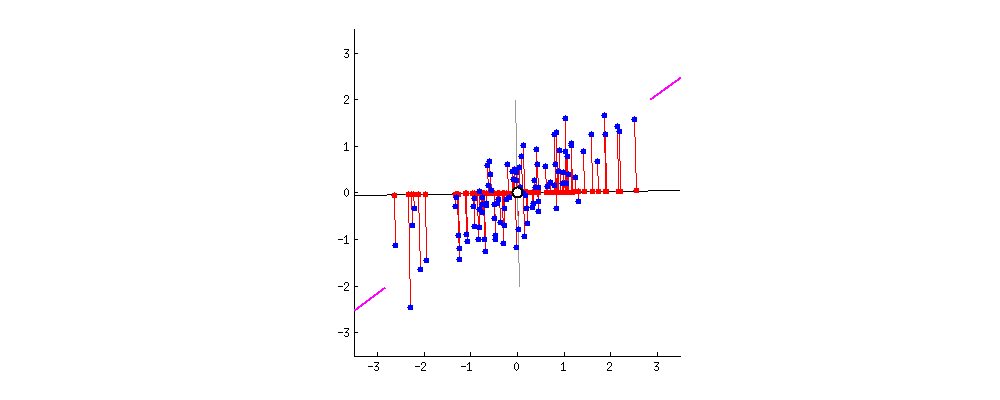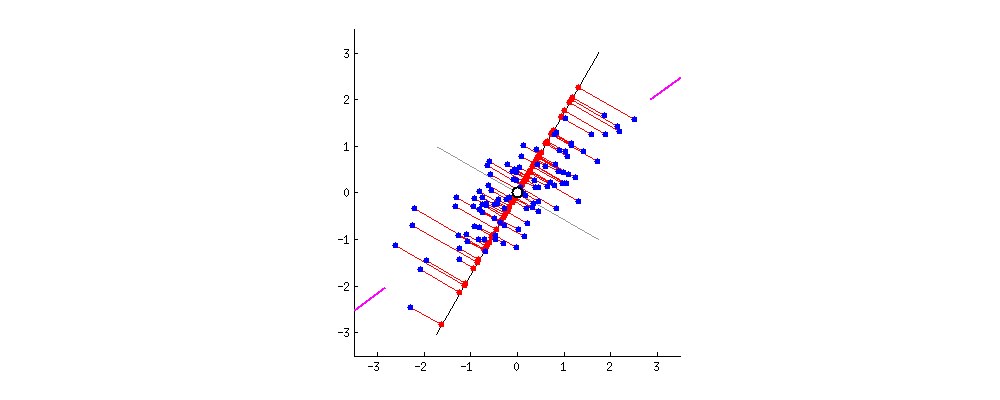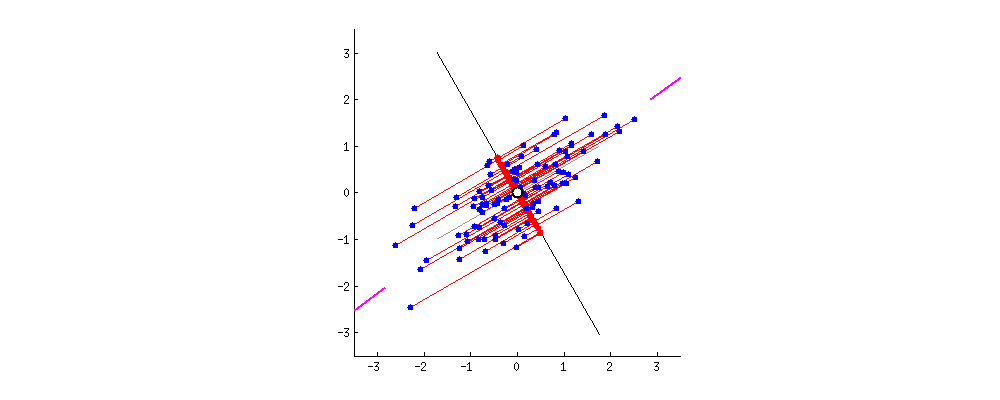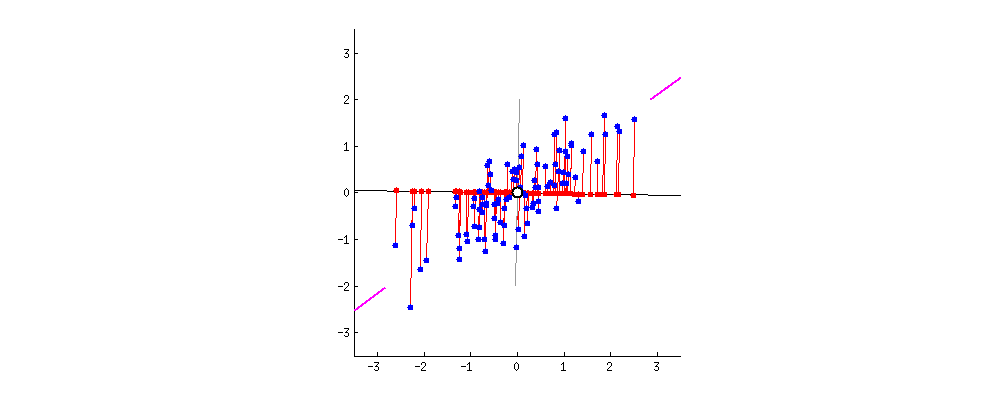
The Meaning of the Best line
Principal Component 1 (PC1): maximizes the variance of the projected points.
-
PC1 is the line in the Eng-Math space that is closest to the \(n\) observations
- PC1 minimizes the sum of squared distances between the data points and the PC1.
PC1 is the best 1D representation of the 2D data
The Meaning of the Best line
PC1 and PC2
- The data points are also spread out a little above and below the PC1.
- There are some variation that is not explained by the PC1.
- Find the second PC, PC2, that
- explains the remaining variation
- is the line through the origin and perpendicular to PC1.
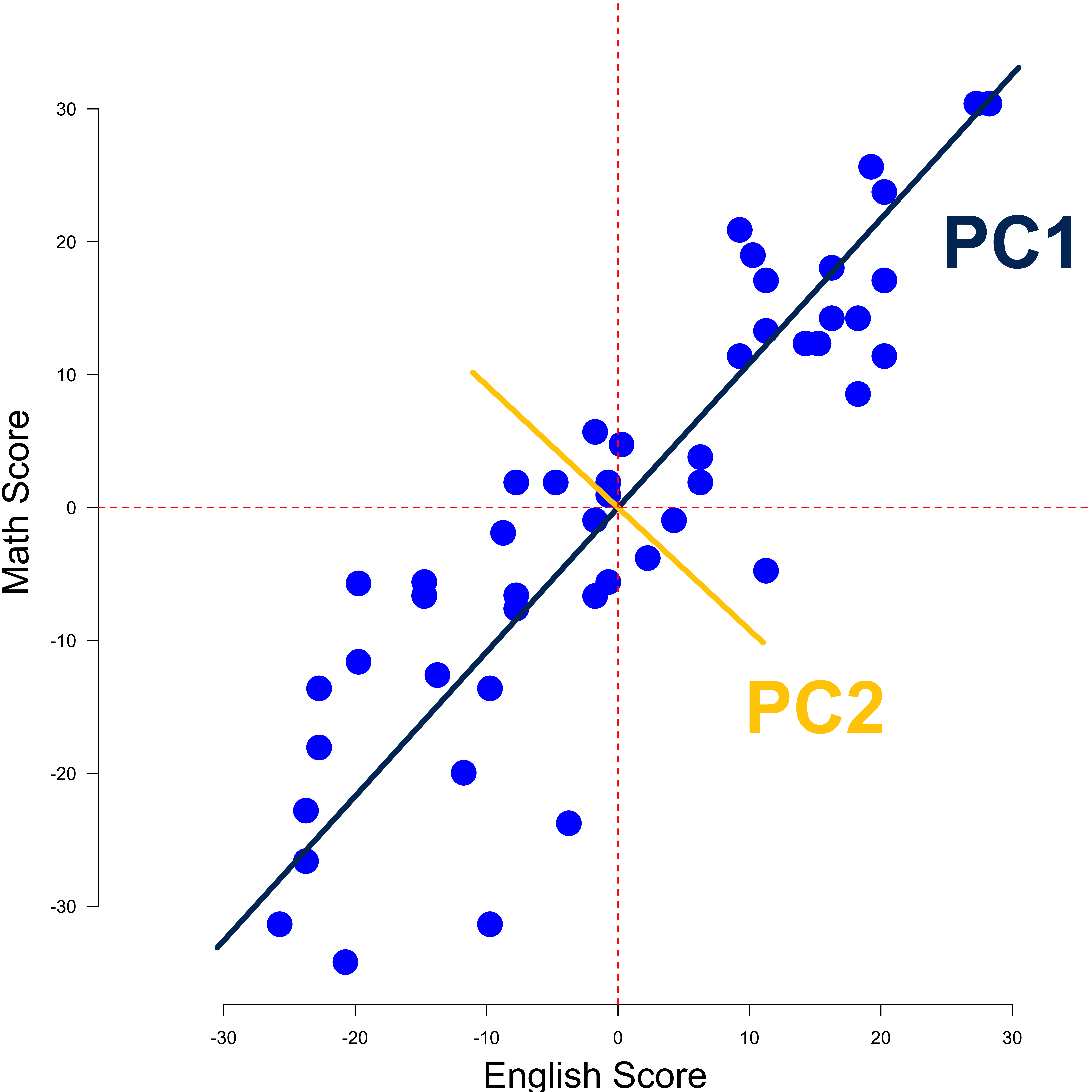
Linear Combinations
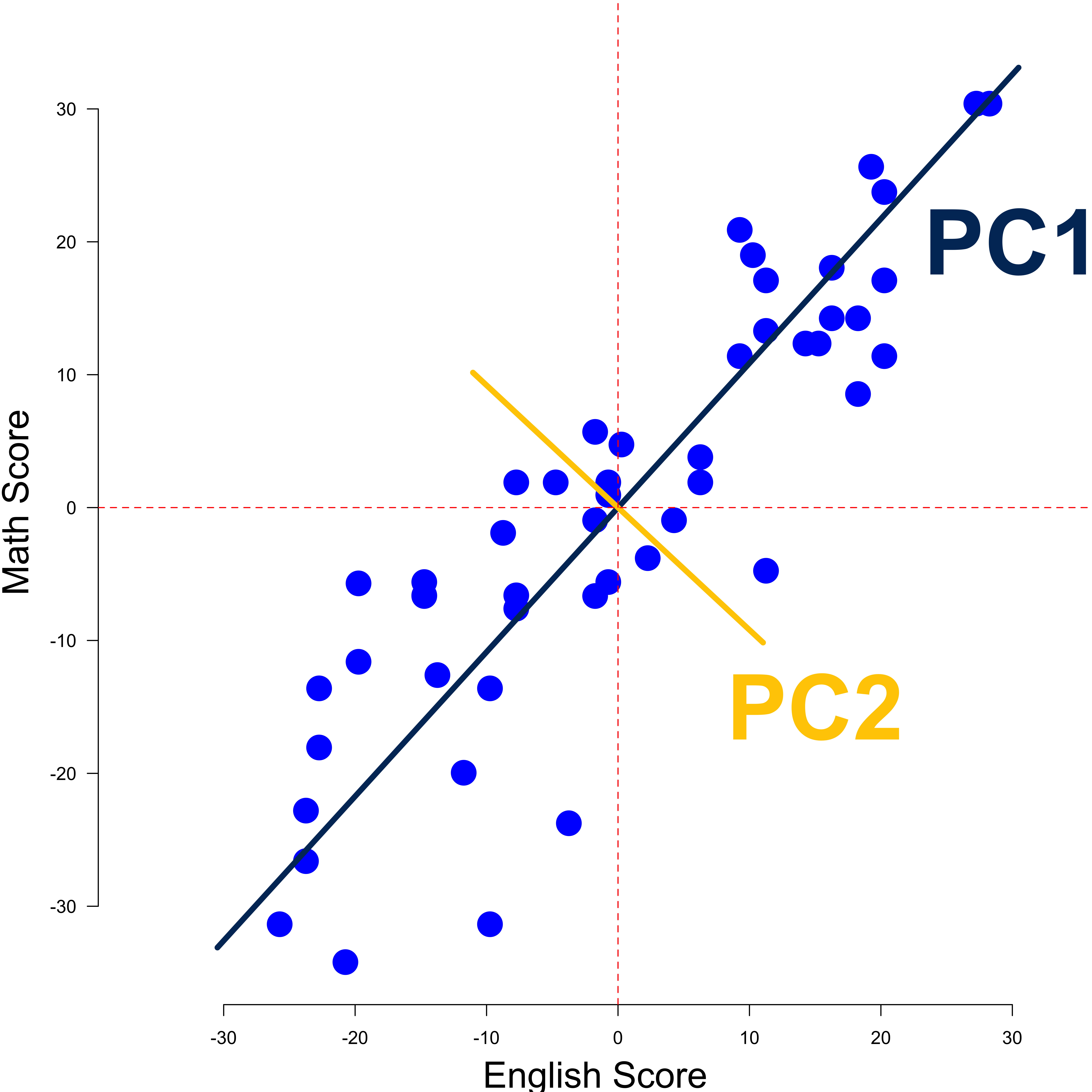
- PC1 = 0.68 \(\times\) English \(+\) 0.74 \(\times\) Math
- PC2 = 0.74 \(\times\) English \(-\) 0.68 \(\times\) Math
- PC1 is like an overall intelligence index as it is a weighted average combining verbal and quantitative abilities.
- PC2 accounts for individual difference in English and Math scores.
- \(0.68^2 + 0.74^2 = 1\) (Pythagorean theorem)
- The combination weights 0.68, 0.74, etc are called PC loadings.
Rotate Everything so that PC1 is Horizontal
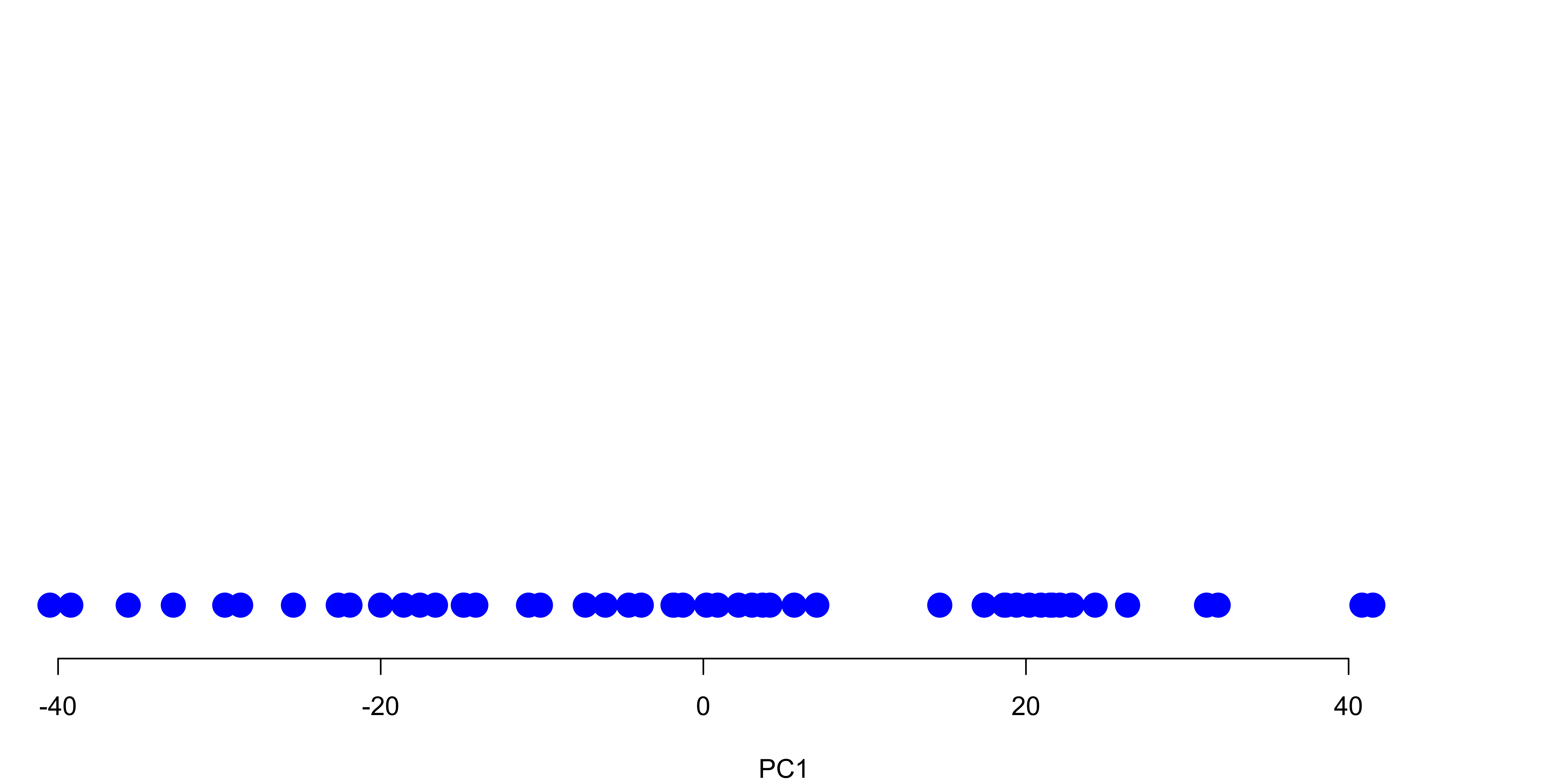
1D representation
- PC1 is our 1D number line that explains the most variation contained in 2D data using a 1D line.
- Points on the PC1 are the projected points of data onto PC1.
2D representation
- The new coordinates PC1 and PC2 are ordered by variation size of the English and Math scores
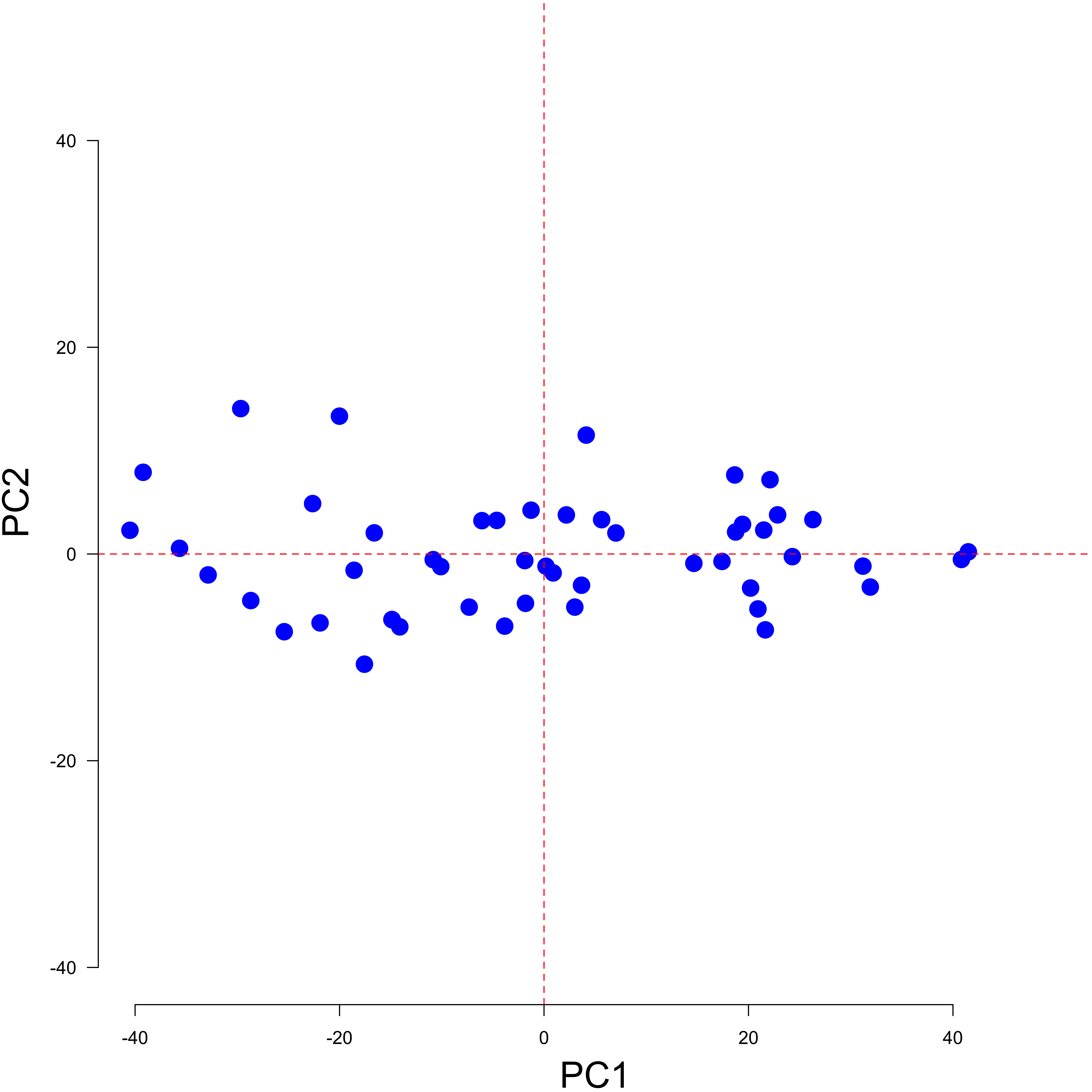
Variation
If the variation for PC1 is \(17\) and the variation for PC2 is \(2\), the total variation presented in the data is \(17+2=19\).
PC1 accounts for \(17/19 = 89\%\) of the total variation, and PC2 accounts for \(2/19 = 11\%\) of the total variation.
How about 3 or More Variables?
- PC1 spans the direction of the most variation
- PC2 spans the direction of the 2nd most variation
- PC3 spans the direction of the 3rd most variation
- PC4 spans the direction of the 4th most variation
- If we have \(n\) observations and \(p\) variables (dimensions), there are at most \(\min(n - 1, p)\) PCs.
US Arrest Data in 1973
Murder Assault UrbanPop Rape
Alabama 13.2 236 58 21
Alaska 10.0 263 48 44
Arizona 8.1 294 80 31
Arkansas 8.8 190 50 20
California 9.0 276 91 41
Colorado 7.9 204 78 39
Connecticut 3.3 110 77 11
Delaware 5.9 238 72 16
Florida 15.4 335 80 32
Georgia 17.4 211 60 26
Hawaii 5.3 46 83 20
Idaho 2.6 120 54 14
Illinois 10.4 249 83 24
Indiana 7.2 113 65 21
Iowa 2.2 56 57 11
Kansas 6.0 115 66 18PC Loading Vectors on USArrests
pca_output <- prcomp(USArrests, scale = TRUE)
## rotation matrix provides PC loadings
(pca_output$rotation <- -pca_output$rotation) PC1 PC2 PC3 PC4
Murder 0.54 0.42 -0.34 -0.649
Assault 0.58 0.19 -0.27 0.743
UrbanPop 0.28 -0.87 -0.38 -0.134
Rape 0.54 -0.17 0.82 -0.089- PCs are unique up to a sign change, so
-pca_output$rotationgives us the same PCs aspca_output$rotationdoes. The sign just change the direction, not the angle.
\(\text{PC1} = 0.54 \times \text{Murder} + 0.58 \times \text{Assault} + 0.28 \times \text{UrbanPop} + 0.54 \times \text{Rape}\)
\(\text{PC2} = 0.42 \times \text{Murder} + 0.19 \times \text{Assault} - 0.87 \times \text{UrbanPop} - 0.17 \times \text{Rape}\)
- We have 4 PCs because \(\min(n-1, p) = \min(50-1, 4) = 4\).
PC Scores
- The value of the rotated data, the data values of each PC are stored in
pca_output$x
PC1 PC2 PC3 PC4
Alabama 0.98 1.12 -0.44 -0.15
Alaska 1.93 1.06 2.02 0.43
Arizona 1.75 -0.74 0.05 0.83
Arkansas -0.14 1.11 0.11 0.18
California 2.50 -1.53 0.59 0.34
Colorado 1.50 -0.98 1.08 0.00
Connecticut -1.34 -1.08 -0.64 0.12
Delaware 0.05 -0.32 -0.71 0.87
Florida 2.98 0.04 -0.57 0.10
Georgia 1.62 1.27 -0.34 -1.07
Hawaii -0.90 -1.55 0.05 -0.89
Idaho -1.62 0.21 0.26 0.49
Illinois 1.37 -0.67 -0.67 0.12
Indiana -0.50 -0.15 0.23 -0.42
Iowa -2.23 -0.10 0.16 -0.02
Kansas -0.79 -0.27 0.03 -0.20Interpretation of PCs
PC1 PC2 PC3 PC4
Murder 0.54 0.42 -0.34 -0.649
Assault 0.58 0.19 -0.27 0.743
UrbanPop 0.28 -0.87 -0.38 -0.134
Rape 0.54 -0.17 0.82 -0.089- PCs are less interpretable than original features.
- The first loading vector places approximately equal weight on
Assualt,MurderandRape, with much less weights onUrbanPop. - PC1 roughly corresponds to a overall serious crime rate.
- The second loading vector places most of its weight on
UrbanPop, and much less weight on the other 3 features. - PC2 roughly corresponds to the level of urbanization.
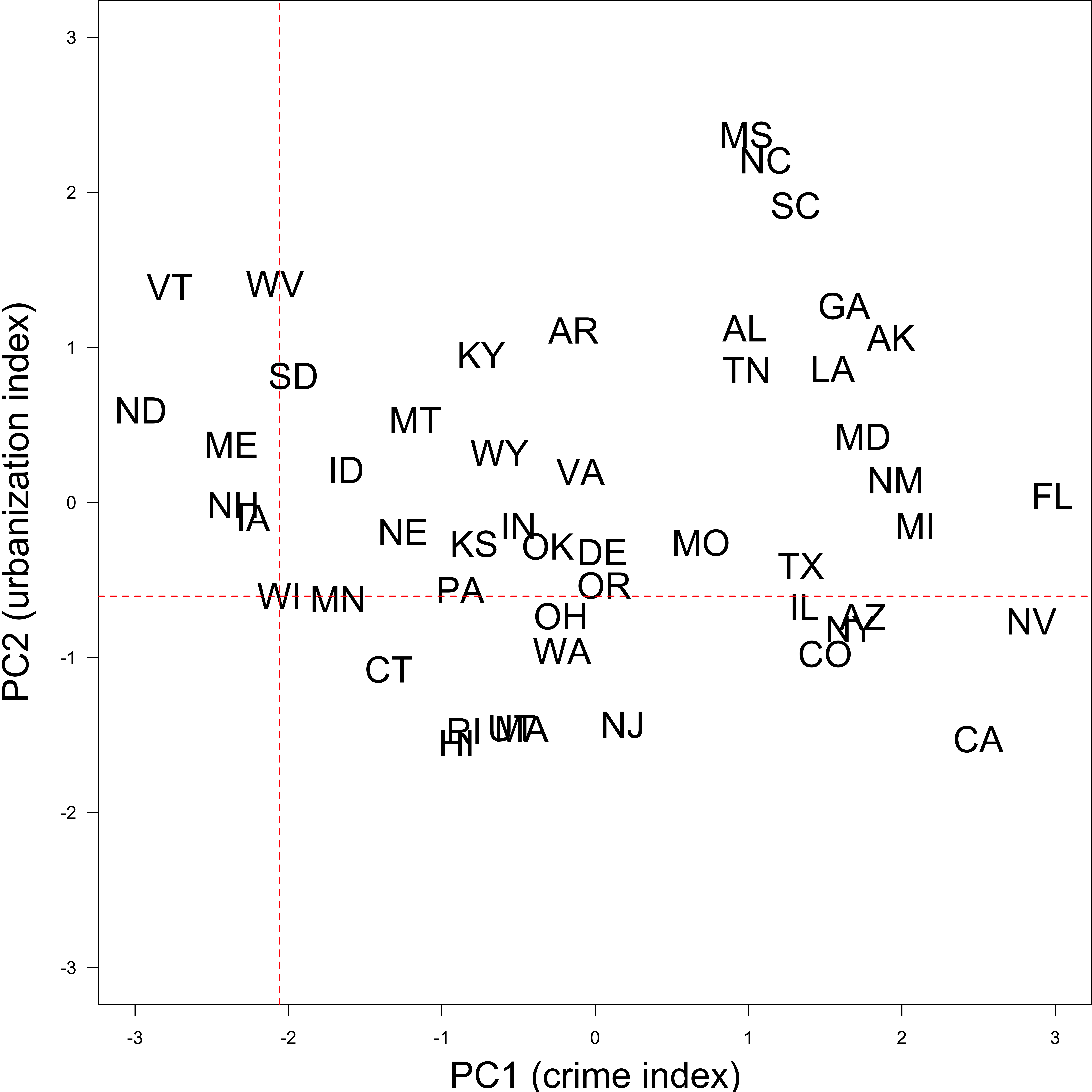
2D Representation of the 4D data
PC1 PC2 PC3 PC4
Wisconsin -2.06 -0.61 -0.14 -0.18
Wyoming -0.62 0.32 -0.24 0.16Higher value of PC1 means higher crime rates (roughly).
Higher value of PC2 means higher level of urbanization (roughly).
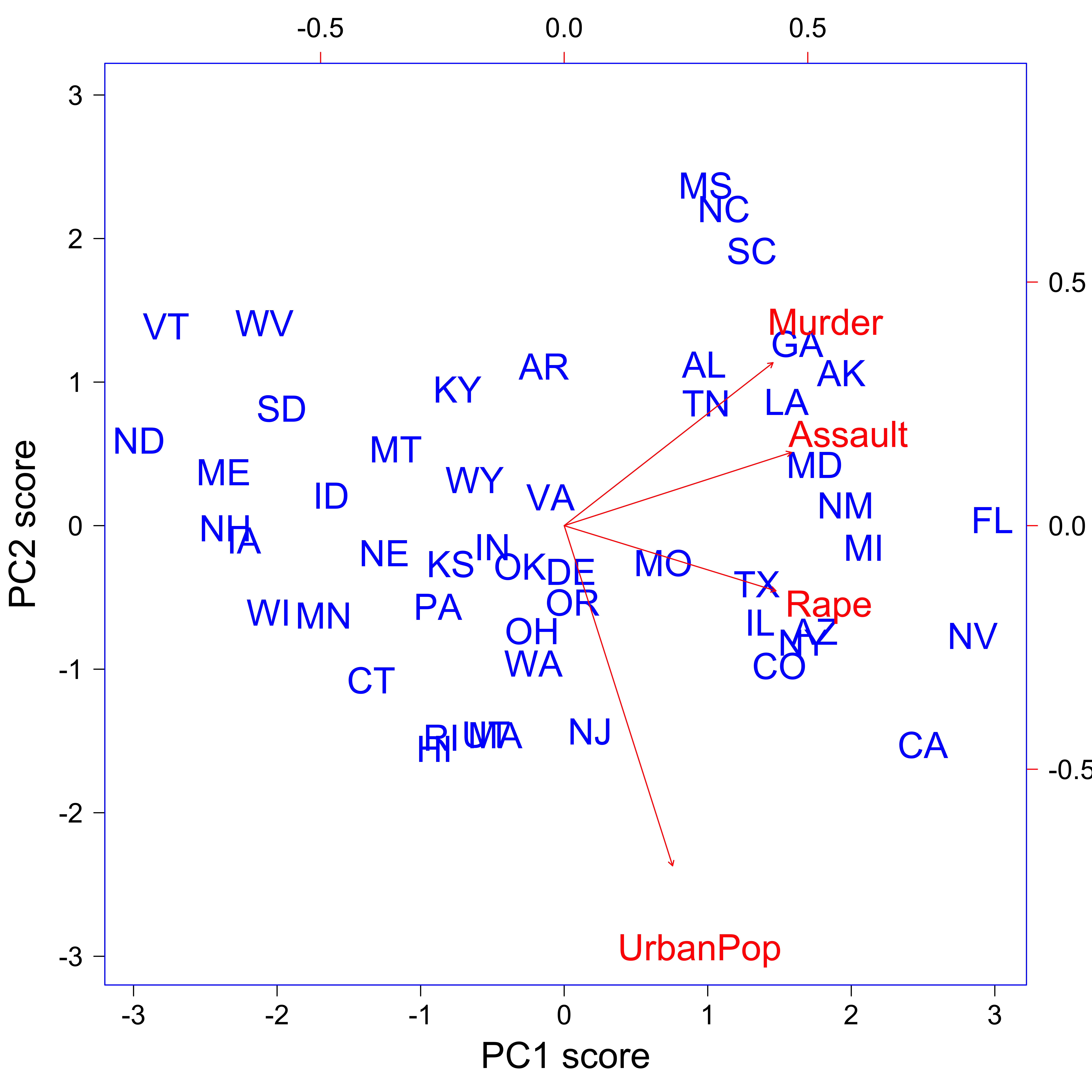
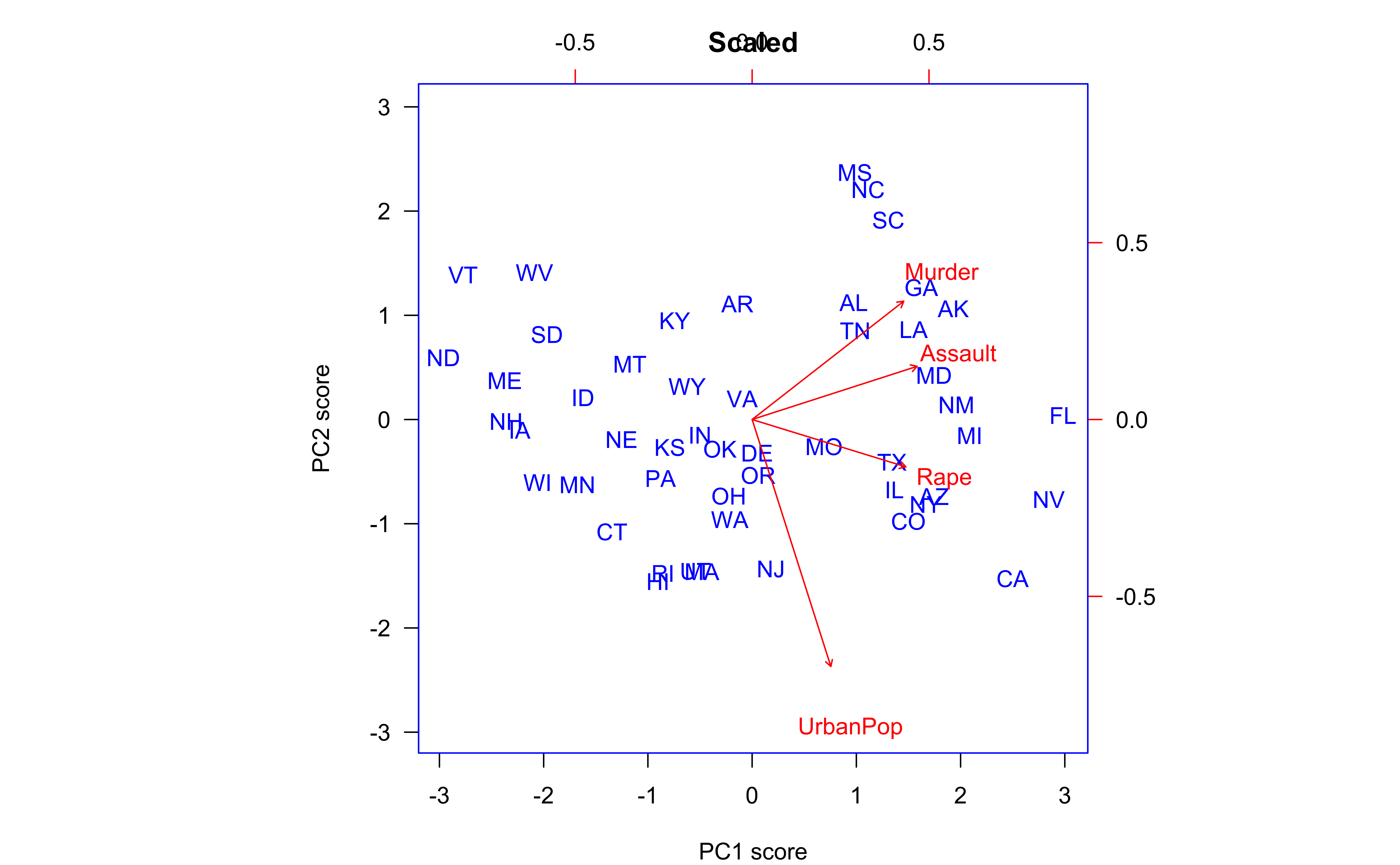
2D Representation of the 4D data: biplot
- Top axis: PC1 loadings
- Right axis: PC2 loadings
- Red arrows: PC1 and PC2 loading vector, e.g., (0.28, -0.87) for
UrbanPop. - Crime-related variables (
Assualt,MurderandRape) are located close to each other. -
UrbanPopis far from the other three. -
Assualt,MurderandRapeare more correlated, andUrbanPopis less correlated with the other three.
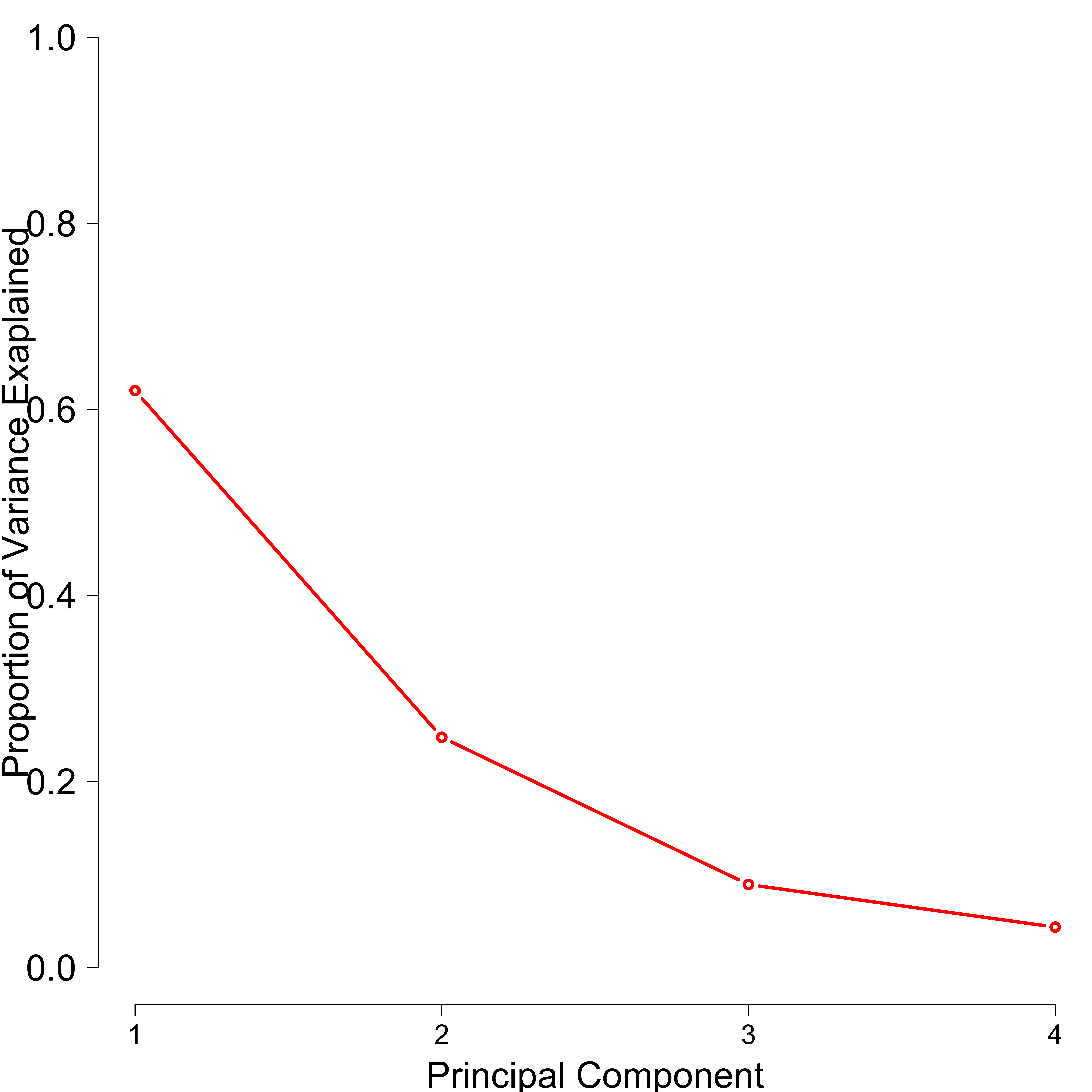
Proportion of Variance Explained
[1] 2.48 0.99 0.36 0.17[1] 0.620 0.247 0.089 0.043- PC1 explains \(62\%\) of the variations in the data, and PC2 explains \(24.7\%\) of the variance.
- PC1 and PC2 explain about \(87\%\) of the variance, and the last two PCs explain only \(13\%\).
- 2D plot provides pretty accurate summary of the data.
Scree Plot
Look for a point at which the proportion of variance explained by each subsequent PC drops off.
Mathematics of PCA
Principal Components
- The \(k\)th PC is
\[Z_k = \phi_{1k}X_1 + \phi_{2k}X_2 + \dots + \phi_{pk}X_p,\] where \(\sum_{j=1}^p\phi_{jk}^2=1\).
\((\phi_{1k}, \phi_{2k}, \dots, \phi_{pk})'\) is the PC loading vector.
The PC1 loading vector solves
\[\max_{\phi_{11}, \phi_{21}, \dots, \phi_{p1}} \left\{ \frac{1}{n}\sum_{i=1}^n z_{i1}^2\right\} = \left\{ \frac{1}{n}\sum_{i=1}^n \left( \sum_{j=1}^p \phi_{j1} x_{ij}\right)^2\right\} \quad \text{s.t.} \quad \sum_{j=1}^p\phi_{j1}^2 = 1\]
Maximize the sample variance of the projected points, or the scores \(z_{11}, z_{21}, \dots, z_{n1}\).
The PC loading vector defines a direction in feature space along which the data vary the most.
Principal Components
For \(k\)th PC, \(k > 1\),
\[\max_{\phi_{1k}, \phi_{2k}, \dots, \phi_{pk}} \left\{ \frac{1}{n}\sum_{i=1}^n z_{ik}^2\right\} = \left\{ \frac{1}{n}\sum_{i=1}^n \left( \sum_{j=1}^p \phi_{jk} x_{ij}\right)^2\right\} \quad \text{s.t.} \quad \sum_{j=1}^p\phi_{jk}^2 = 1, \text{ and } {\mathbf{z}_m}'\mathbf{z}_k = 0, \, m = 1, \dots, k-1\] where
\(\mathbf{z}_l = (z_{1l}, z_{2l}, \dots, z_{nl})'\)
Low-Rank Approximation
PCs provide low-dimensional planes that are closest to the observations.
\(x_{ij} \approx \sum_{m=1}^M z_{im}\phi_{jm}\) with equality when \(M = \min(n-1, p)\)
\[(z_{im}, \phi_{jm}) = \mathop{\mathrm{arg\,min}}_{a_{im}, b_{jm}} \left\{ \sum_{j=1}^p\sum_{i=1}^n\left( x_{ij} - \sum_{m=1}^Ma_{im}b_{jm}\right)^2\right\}\]
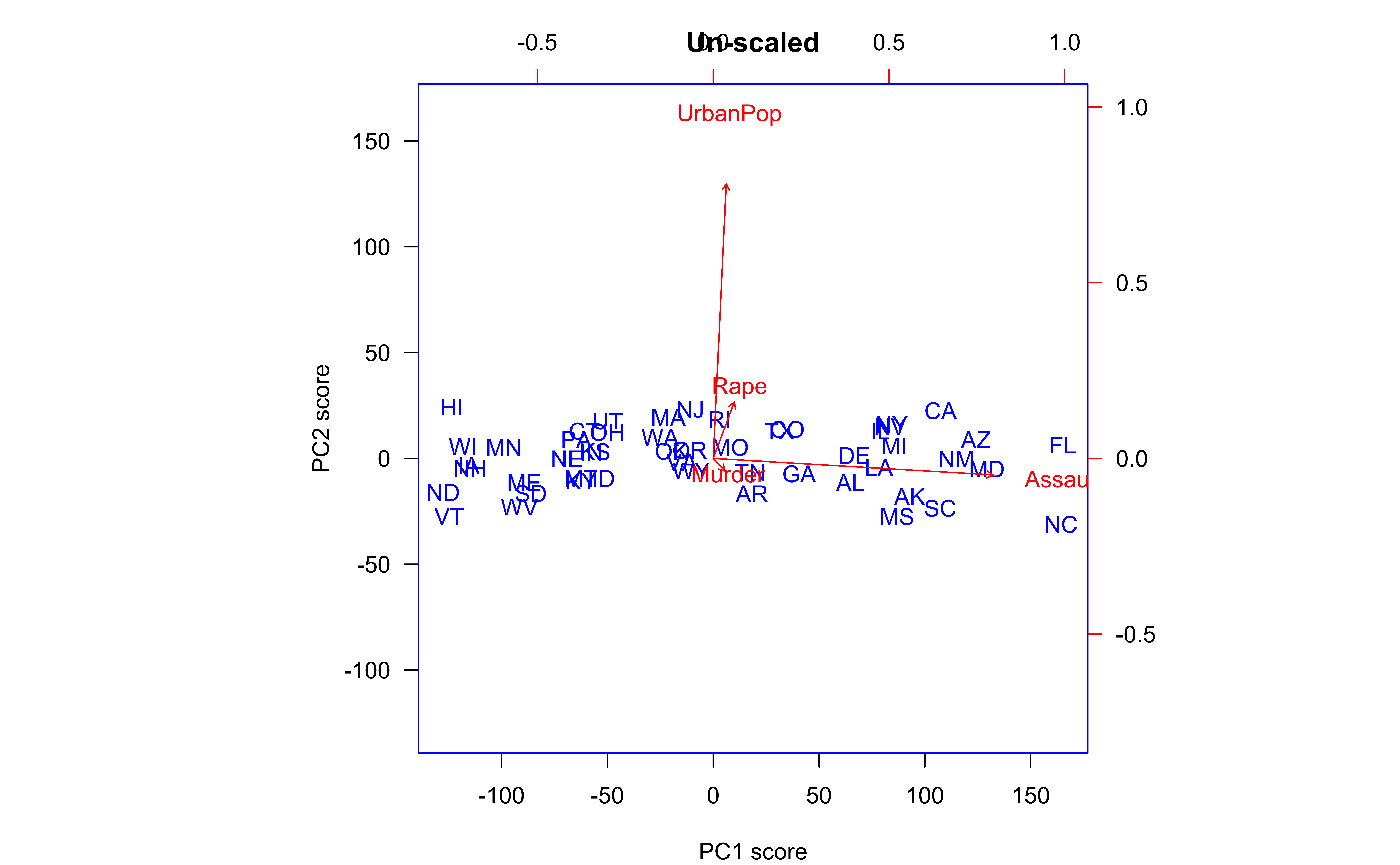
Scaling the Variables
If we perform PCA on the unscaled variables, PC1 loading vector will have a large loading for
Assault.When all the variables are of the same type, no need to scale the variables.
PCA and SVD
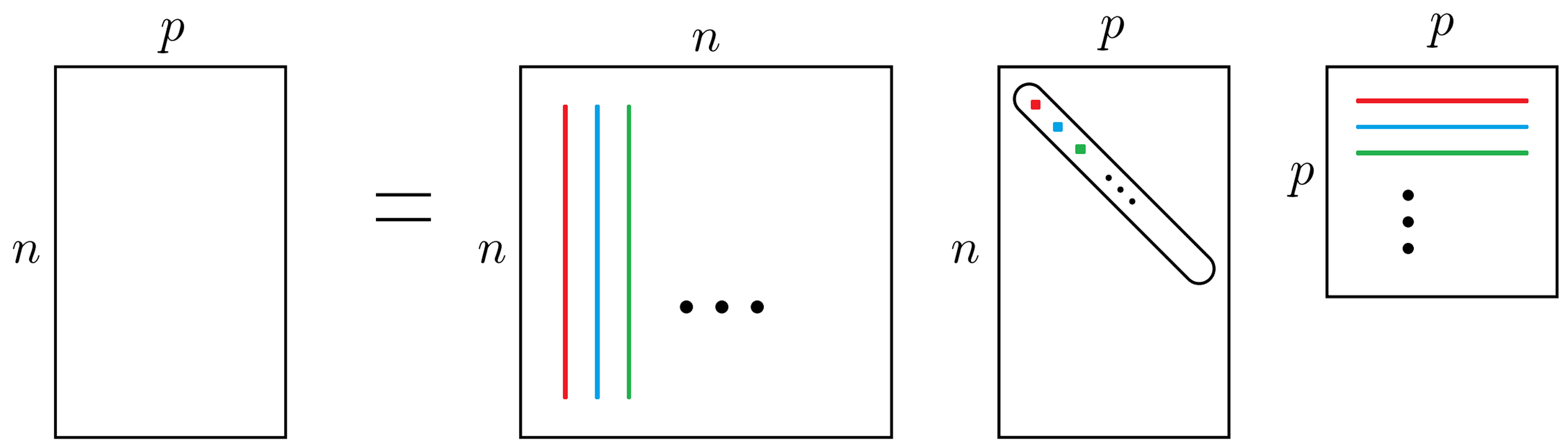
PCA is equivalent to singular value decomposition (SVD) of \(\mathbf{X}\)
\[\mathbf{X}_{n\times p} = \mathbf{U}_{n \times n} \mathbf{D}_{n \times p} \mathbf{V}'_{p \times p}\] where \(\mathbf{U}\) and \(\mathbf{V}\) are orthogonal matrices, and \(\mathbf{D}\) is diagonal with diagonal elements singular values \(d_1 \ge d_2 \ge \cdots \ge d_p\).
PC Scores and SVD
\({\bf Z}_{n \times p} = [{\bf z}_1 \, {\bf z}_2 \, \cdots \, {\bf z}_p] = \mathbf{X}\mathbf{V}\)
The \(j\)th PC is the \(j\)th column of \({\bf Z}\) given by \({\bf z}_j = (z_{1j}, z_{2j}, \dots, z_{nj})' = {\bf Xv}_j\).
Project \(\mathbf{X}\) onto the space spanned by \(\mathbf{v}_j\)s
\(\mathbf{v}_j\)s are loading vectors.
\({\bf Z}_{n \times p} = \mathbf{U}\mathbf{D}\)
\({\bf z}_j = d_j\mathbf{u}_j\).
\(\mathbf{u}_j\)s are the unit PC vectors, and \(d_j\) controls the variation along the \(\mathbf{u}_j\) direction.
Low-Rank Approximation and SVD
\({\mathbf{X}} = {\mathbf{U}}{\mathbf{D}}{\mathbf{V}}'\)
\(\mathbf{A}= \mathbf{U}_{n\times M}\mathbf{D}_{M \times M}\) and \(\mathbf{B}' = \mathbf{V}_{M\times p}'\) are the minimizer of
\[ \min_{\mathbf{A}\in \mathbf{R}^{n \times M}, \mathbf{B}\in \mathbf{R}^{p \times M}} \|\mathbf{X}- \mathbf{A}\mathbf{B}' \|\] where
- \(\mathbf{U}_{n\times M}\) is \(\mathbf{U}\) with the first \(M\) columns
- \(\mathbf{D}_{M \times M}\) is \(\mathbf{D}\) of the first \(M\) rows and columns
- \(\mathbf{V}_{M\times p}'\) is \(\mathbf{V}'\) with the first \(M\) rows
\(x_{ij} = \sum_{m = 1}^p d_mu_{im}v_{jm}\)
\(x_{ij} \approx \sum_{m = 1}^M d_mu_{im}v_{jm}\)
PCA and Eigendecomposition
PCA is equivalent to eigendecomposition of \(\mathbf{X}'\mathbf{X}\) or \(\boldsymbol \Sigma= \text{Cov}(\mathbf{X}) = \dfrac{\mathbf{X}'\mathbf{X}}{n-1}\)1, the covariance matrix of \(\mathbf{X}\).
\[\mathbf{X}'\mathbf{X}= \mathbf{V}\mathbf{D}^2\mathbf{V}' = d_1^2\mathbf{v}_1\mathbf{v}_1' + \dots + d_p^2\mathbf{v}_p\mathbf{v}_p'\]
Total variation: \(\sum_{j=1}^p \text{Var}(\mathbf{x}_j) = \frac{1}{n-1}\sum_{j=1}^pd_j^2 = p\)
Variation of \(m\)th PC: \(\text{Var}(\mathbf{z}_m) = \frac{d_m^2}{n-1}\)
Transformed OLS Regression
Transform \({\bf y = X\boldsymbol \beta+ \boldsymbol \epsilon}\) into \[{\bf y = XVV'\boldsymbol \beta+ \boldsymbol \epsilon= Z\boldsymbol \alpha} + \boldsymbol \epsilon\] where \(\mathbf{Z}= \mathbf{X}\mathbf{V}\) and \(\boldsymbol \alpha= \mathbf{V}'\boldsymbol \beta\), or \(\boldsymbol \beta= \mathbf{V}\boldsymbol \alpha\).
The least- squares estimator \(\hat{\boldsymbol \alpha} = (\mathbf{Z}'\mathbf{Z})^{-1}\mathbf{Z}'\mathbf{y}= \mathbf{D}^{-2}\mathbf{Z}'\mathbf{y}\)
\(\text{Var}\left(\hat{\boldsymbol \alpha} \right) = \sigma^2 (\mathbf{Z}'\mathbf{Z})^{-1} = \sigma^2 \mathbf{D}^{-2}\)
A small \(d_j\) means that the variance of \(\alpha_j\) will be large.
-
The PC regression combats multicollinearity by using less PCs \((m \ll p)\) in the model.
- The PCs corresponding to tiny \(d_j\)s (huge variance) are removed and least-squares is applied to the remaining PCs.
Principal Component Regression (PCR)
- Keep \(m < p\) PCs with the largest \(m\) singular values \(d_1, d_2, \dots, d_m\).
- The least- squares estimator is \[\hat{\boldsymbol \alpha}_m = ({\bf Z} _m ' {\bf Z} _m) ^ {-1} {\bf Z}_m ' {\bf y} = \mathbf{D}_m ^ {-2} {\bf Z}_m '{\bf y}\] where \({\bf Z}_m = [{\bf z}_1 \, {\bf z}_2 \, \cdots \, {\bf z}_m]\) and \({\bf \mathbf{D}}_m = \text{diag}(d_1, d_2, \dots, d_m)\)
- \({\bf b}_{pc} = {\bf V}_m\hat{\boldsymbol \alpha}_m = [{\bf v}_1 \, {\bf v}_2 \, \cdots \, {\bf v}_m] \begin{bmatrix} \hat{\alpha}_1 \\ \hat{\alpha}_2 \\ \vdots \\ \hat{\alpha}_m \end{bmatrix}\)
- \({\bf b}_{pc}\) is biased but has smaller variance than \({\bf b}\).
Principal Component Regression
- PCR performs well when the directions in which \(X_1, \dots ,X_p\) show the most variation (the first few PCs) are the directions that are associated with \(Y\).
- Deleting PCs does not imply deletion of any of original regressors. The selected \(m\) PCs contain information provided by all \(k\) original regressors. (Not a feature selection method)
- PCR and ridge regression are closely related. Ridge regression is a continuous version of PCR.
pls::pcr()
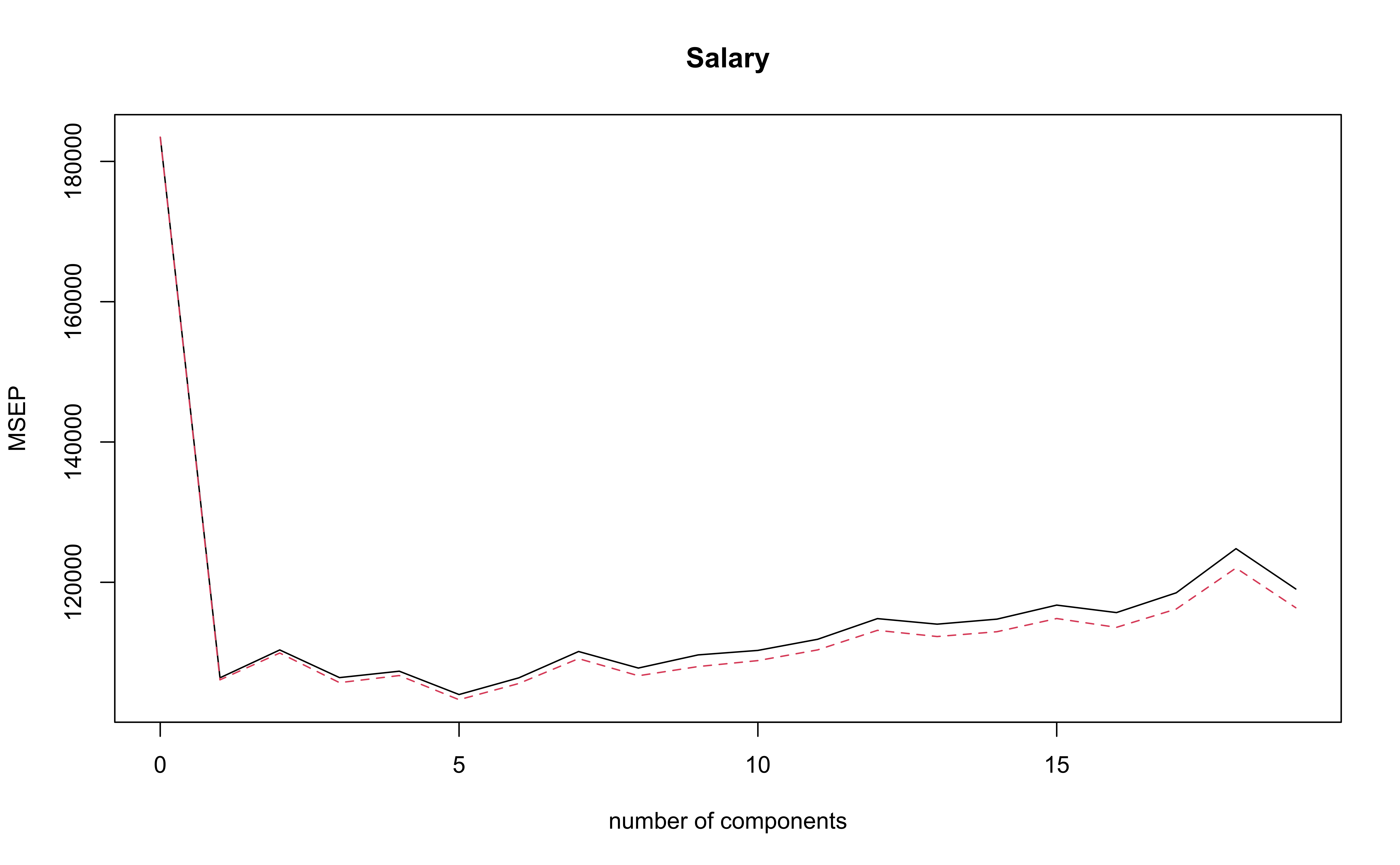
pls::pcr()
[1] 142812Data: X dimension: 263 19
Y dimension: 263 1
Fit method: svdpc
Number of components considered: 5
TRAINING: % variance explained
1 comps 2 comps 3 comps 4 comps 5 comps
X 38.31 60.16 70.84 79.03 84.29
y 40.63 41.58 42.17 43.22 44.90Other Dimension Reduction (Latent Variable) Methods
Kernel Principal Component Analysis https://ml-explained.com/blog/kernel-pca-explained
Probabilistic PCA
Factor Analysis
Autoencoders
t-SNE